 Written by: Genya Gorshtein, MSc
Written by: Genya Gorshtein, MSc
Published: September 3, 2021
Updated: October 29, 2024
Protein sequencing involves determining the precise order of amino acids in a protein or peptide, which allows for the identification and detailed characterization of the protein.
Understanding proteins at a fundamental level—by knowing their exact amino acid sequence—is essential for unlocking insights into how they function, interact, and contribute to both health and disease.
Applications of Protein Sequencing
Knowing the sequence is crucial for understanding a protein’s structure, function, and potential modifications that may affect its activity. As such, protein sequencing is an invaluable tool across various scientific disciplines and applications.
Antibody Discovery
Antibodies are highly variable and diverse, with an estimated 1018 different variants originating from three immunoglobulin loci in the human genome. This vast diversity makes it challenging to discover antibodies using only DNA or genome databases. Sequencing is essential for identifying and characterizing antibodies, particularly for therapeutic and diagnostic applications.
Case Study: De Novo Protein Sequencing of Antibodies for Identification of Neutralizing Antibodies in Human Plasma
Biomarker Discovery
Protein sequencing is essential for identifying biomarkers—proteins whose presence or alteration indicates a disease state. By sequencing the proteins from patient samples, researchers can detect these disease-associated changes with high precision, improving diagnostic accuracy and enabling personalized medicine.
Therapeutic Development
In the pharmaceutical industry, sequencing proteins helps identify and verify the structure of therapeutic proteins to ensure their safety and efficacy. Additionally, it supports the development of biosimilar drugs, which are highly similar versions of biologic drugs.
Reagent Validation
Reagent validation using protein sequencing ensures the accuracy and reliability of biologic-based research tools, such as antibodies. Researchers can confirm the identity and integrity of reagents, preventing issues like batch-to-batch variation, cross-reactivity or contamination. This process enhances the reproducibility of experiments and increases confidence in scientific results.
Case Study: Generation and Diversification of Recombinant Monoclonal Antibodies
Structural Biology
Knowledge of the amino acid sequence is highly beneficial and often critical for structural biology. Techniques such as cryo-electron microscopy, X-ray crystallography, and nuclear magnetic resonance (NMR) rely on the amino acid sequence for resolving and interpreting the 3D structure of proteins.
Studying Protein Variants
Proteins are subject to variations due to mutations, alternative splicing, or proteolytic processing following translation. These modifications are not encoded for or identified through DNA sequencing, and thereby requires protein sequencing to analyze and identify these protein variants.
Identifying Post-Translational Modifications
After proteins are translated, they undergo post-translational modifications (PTMs). PTMS including phosphorylation, glycosylation, acetylation, or methylation alter a protein’s activity, stability, and interactions with other molecules. Mass spectrometry-based protein sequencing provides the ability to detect and map PTMs.
Methods for Protein Sequencing
While protein and peptide sequencing methods have historically lagged behind DNA sequencing, advancements in new technologies have significantly improved the field, enabling more comprehensive proteomic studies.
Edman Degradation
Edman degradation is one of the earliest approaches to protein sequencing, involving the stepwise degradation of peptides to derive the order of amino acid residues.
In this process, the N-terminal amino acid of the protein is labeled and then cleaved off without disrupting the peptide bonds between the remaining amino acids. The released amino acid is identified through chromatography or electrophoresis and repeated for each successive amino acid. Edman degradation is most effective for shorter peptides, typically up to 50 amino acids.
Mass Spectrometry
The integration of liquid chromatography and mass spectrometry (LC-MS) into protein sequencing workflows has significantly improved accuracy, sensitivity, and throughput.
LC-MS-based techniques allow researchers to directly determine the amino acid sequences of proteins from biological samples, often without requiring prior genetic information. In this process, proteins are digested by protease enzymes, which cleave them at specific sites, generating a variety of peptides (Figure 1). These peptides are then ionized and analyzed by the mass spectrometer, which measures their masses. Using this data, algorithms reconstruct the protein’s amino acid sequence through both database-driven and de novo sequencing approaches. De novo sequencing is particularly valuable when genomic data is unavailable, as it determines the sequence directly from mass spectra without relying on existing databases. LC-MS is especially useful for species without well-characterized genomes or for analyzing complex protein mixtures.
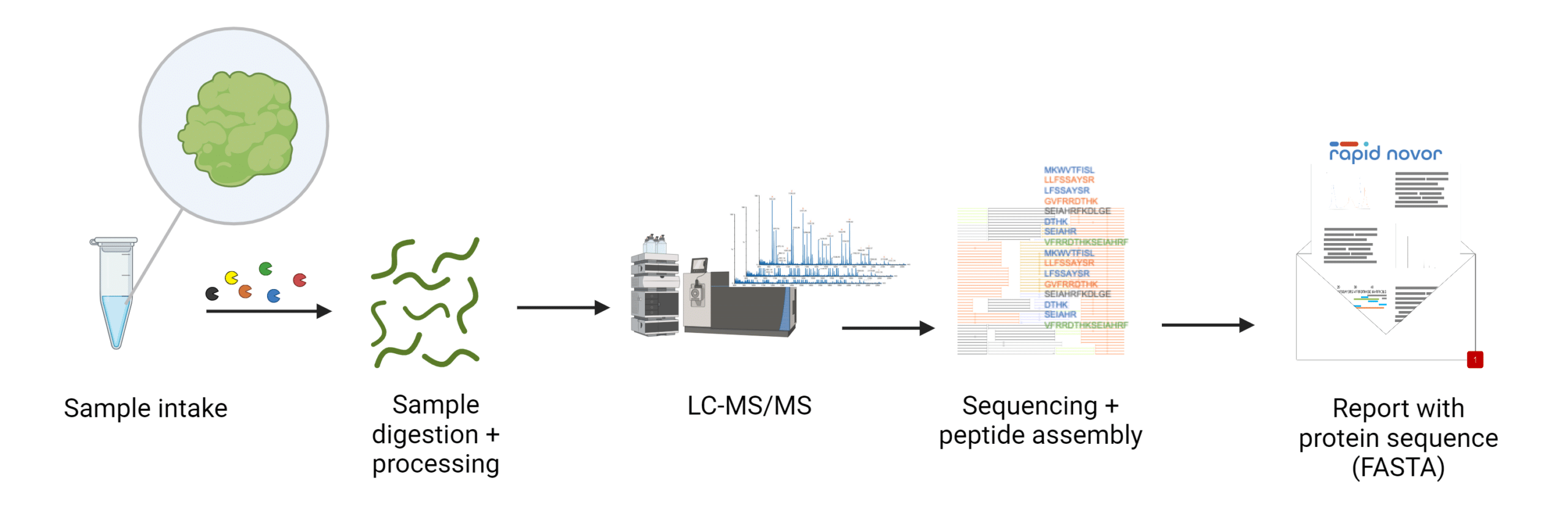
Figure 1. Workflow for LC-MS-based protein sequencing.
Mass Spectrometry-Based Protein Sequencing with Rapid Novor
Rapid Novor is a world leader in protein sequencing, leveraging cutting-edge expertise in mass spectrometry, proteomics, biochemistry, and bioinformatics. REmAb® de novo protein sequencing service delivers the complete protein sequence with unparalleled accuracy and reliability from as little as 100μg of sample. For complex protein mixtures, such as polyclonal antibodies, REpAb® polyclonal antibody sequencing service can accurately sequence individual antibodies directly from purified protein mixtures or immunoserum.
For routine protein sequence confirmation, MATCHmAb™ peptide mapping utilizes LC-MS to offer a fast, reliable solution to verify that the sequence of your produced or procured protein matches the expected sequence. This method can also identify any anomalies for further investigation. Since protein sequence directly influences structure and function, rapid confirmation with MATCHmAb™ serves as an excellent complement to performance validation assays, ensuring the integrity of the protein for downstream applications.
Talk to Our Scientists.
We Have Sequenced 10,000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and developed the first recombinant polyclonal antibody diagnostics.
Talk to Our Scientists.
We Have Sequenced 9000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables timely and reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and ran the first recombinant polyclonal antibody diagnostics

