 Written by Genya Gorshtein, MSc
Written by Genya Gorshtein, MSc
August 10, 2022
General Structure of Antibodies
Antibodies or immunoglobulins (Ig) maintain a common quaternary structure consisting of two identical heavy chains (HCs) and two identical light chains (LCs). The structure is heterotetrameric, with disulfide bridges linking HCs and LCs together to form the canonical immunoglobulin ‘Y’ shape.
Variable domains (V) of heavy and light chains (VH and VL, respectively) give rise to antigen specificity through highly variable amino acid sequences. While the V domain interacts with antigens, the constant domains (C) on heavy and light chains interact with effector proteins and molecules. HCs have three constant regions, in contrast to the one on LCs and as such, are numbered from the N-terminus to the C-terminus (CH1, CH2 and CH3) (Figure 1).
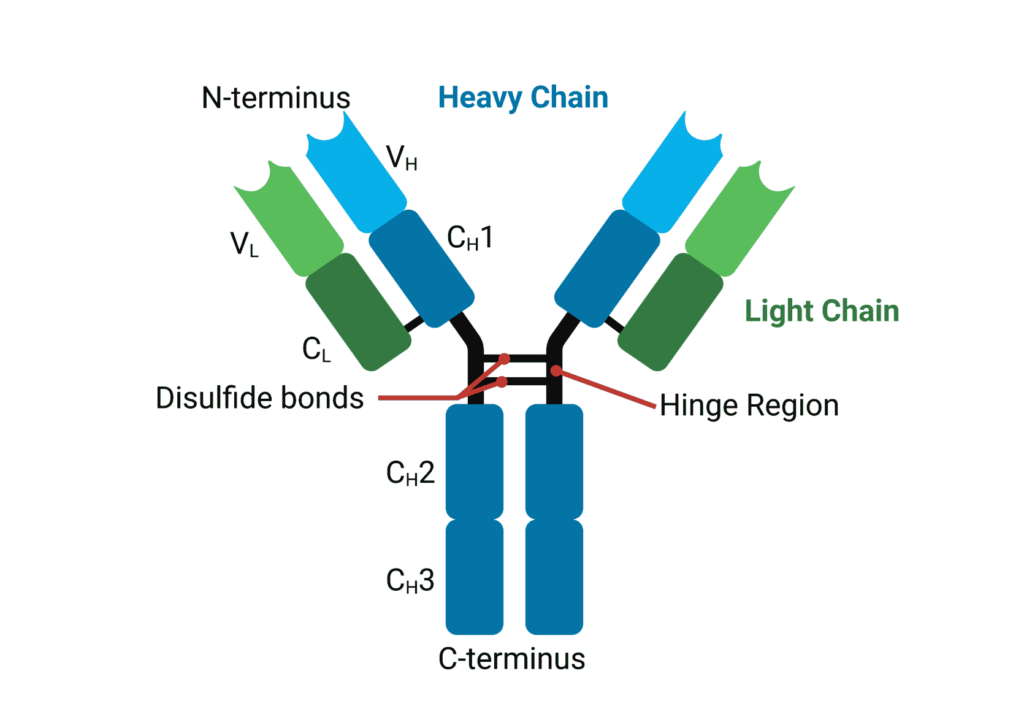
Figure 1. General structure of an immunoglobulin G.
Antibody Heavy Chains
The constant region of the heavy chain confers Ig isotype (also known as class), complement binding, half-life length, and interactions with Fc receptors (FcR). The 5 isotypes of Ig are immunoglobulin M (IgM), immunoglobulin D (IgD), immunoglobulin A (IgA), immunoglobulin E (IgE), and immunoglobulin G (IgG), and are classified according to the type of heavy chain they contain – mu, delta, alpha, epsilon or gamma. IgG is the most abundant Ig with 4 different subclasses in humans (IgG 1, 2, 3, and 4). See our previous article about Ig isotypes.
Antibody Light Chains
In contrast to heavy chains, there are only two types of light chains, lambda (λ) and kappa (κ). Light chain isotypes have no functional differences, however a given antibody will only have κ or λ light chains, but not both. The ratio of κ to λ light chains varies from species to species. In humans, κ to λ ratio is 2:1, whereas in mice it is 20:1. Two additional L-chain types, σ and σ-cart are present in amphibians (only σ), and cartilaginous and bony fish (σ and σ-cart).
Antibody Fab and Fc Fragments
Early knowledge of antibody structure was based on enzymatic digest-based characterization of IgG with papain and pepsin. Papain, a cysteine protease enzyme, cleaves the immunoglobulin into three fragments. Two identical fragments containing the antigen-binding activity are produced – termed the fragment antigen binding, or Fab fragment. This contains the complete light chain (VL and CL) with VH and CH1 of the heavy chain (Figure 2A). The Fc fragment (which stands for Fragment crystallizable) is also produced containing paired CH2 and CH3 domains. Pepsin cleaves immunoglobulins into pFc′ and a dimeric Fab, known as F(ab′)2, which maintains the same binding properties as the intact immunoglobulin without interacting with effector molecules (Figure 2B).
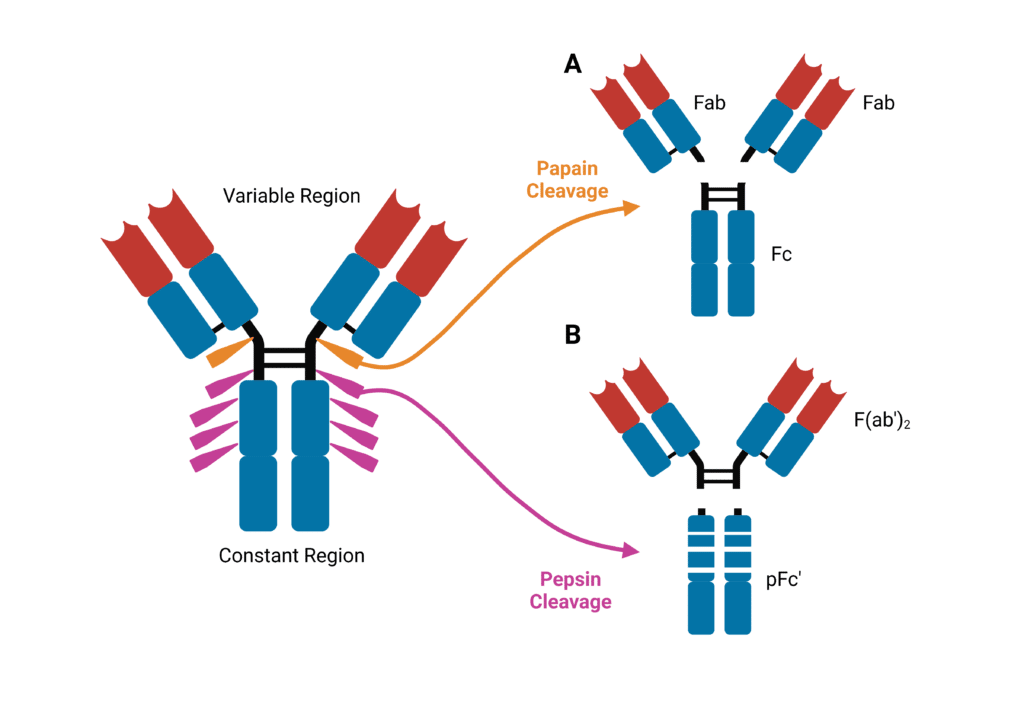
Figure 2. Immunoglobulins are sensitive to cleavage by proteases. (A) Cleavage by papain yields two identical Fab fragments and an Fc fragment. (B) cleavage by pepsin yields a F(ab′)2 and pFc′ fragment.
Hinge Region
Connecting CH1 and CH2 is the hinge region. This region provides a large degree of Fab flexibility, enabling the antibody to bind multiple targets at various distances and interact with effector molecules. The hinge region is particularly susceptible to protease cleavage such as papain and pepsin.
Complementarity Determining Regions and Framework Regions
Within the VH and VL domains are three complementarity-determining regions: CDR-H1, CDR-H2, and CDR-H3 for VH and CDR-L1, CDR-L2, CDR-L3 for VL. These regions are hypervariable in their amino acid sequences, and form distinct loops creating a surface complementary to the antigen. CDR-H3 is the main contributor to antigen recognition due to its sequence diversity, length, and favourable location. The CDR-H3 loop influences loop conformations and antigen binding at the other CDRs, and thereby influences specificity and affinity. Between the CDRs are framework regions (FR) that show less variability and provide structural stability for the domain. These FR regions form a β sheet structure, whereas the CDR loops are located at the outer edge, presenting an antigen-binding site.
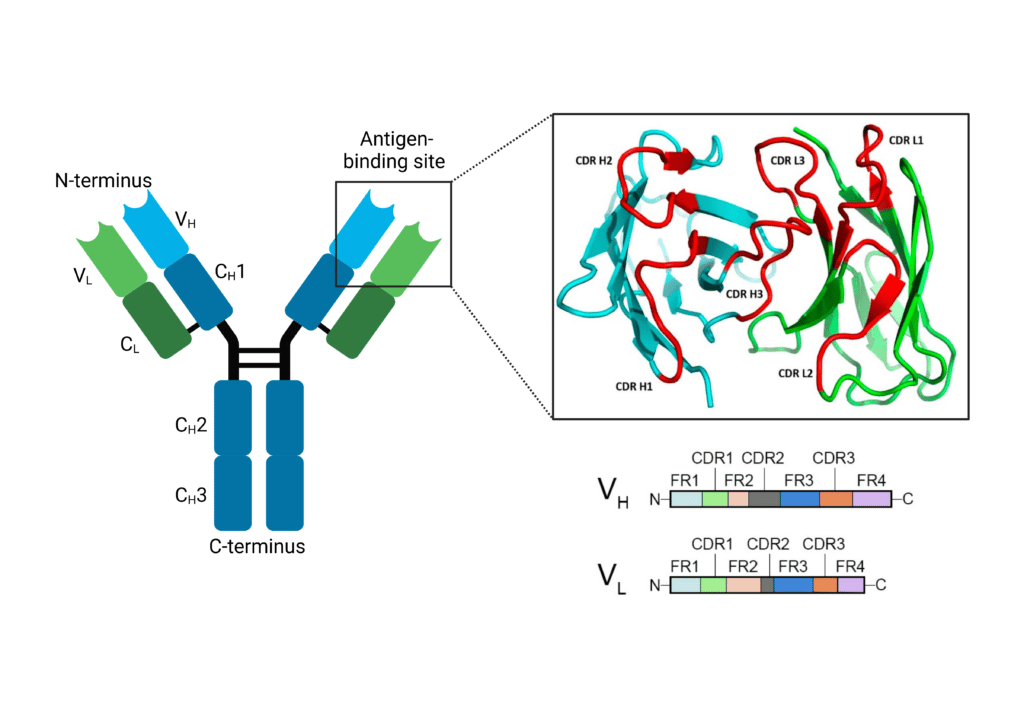
Figure 3. The antigen binding site of an immunoglobulin contains CDR and FR regions. CDR regions (L1-3 and H1-3) make up the antigen binding site on the N-terminus of the immunoglobulin.
Obtaining Antibody Structures With Protein Sequencing and HDX-MS
Rapid Novor provides multiple services that can facilitate the analysis and characterization of antibody sequences and structures de novo protein sequencing services, peptide mapping, and HDX-MS services—to learn more about these technologies, and reach out to our scientists for inquiries.
Talk to Our Scientists.
We Have Sequenced 10,000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and developed the first recombinant polyclonal antibody diagnostics.
Talk to Our Scientists.
We Have Sequenced 9000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables timely and reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and ran the first recombinant polyclonal antibody diagnostics

