March 23, 2021

*Author
#Reviewer
1Rapid Novor, Inc.
Abstract
Mouse monoclonal antibodies (mAbs) are highly attractive for manipulation for therapeutic applications as their manufacturing is relatively easy and well-established compared to mAbs derived from larger animal models. However, they also pose several challenges which limit their use as therapeutic agents. Particularly, their short serum half-life, inability to trigger human effector functions, and their immunogenicity that leads to recognition by the patients’ immune system (i.e., human anti-mouse antibody response). Through protein engineering, murine antibodies have been transformed into structures more similar to human antibodies without sacrificing their binding specificity or affinity, while reducing immunogenicity. In this review, current antibody engineering technologies are explored while discussing how de novo protein sequencing can enhance engineering and characterization efforts for robust mAb therapeutic development.
Key Takeaways
- Thanks to antibody engineering, it has been possible to manipulate antibody structure to generate more robust antibodies while lowering immunogenicity
- However, current methods of generating monoclonal antibodies, and techniques in antibody engineering tend to heavily rely on DNA sequencing methodologies
- Though critical in antibody engineering, DNA manipulation techniques are simply blind to data that protein sequencing can provide
- Thanks to protein sequencing, engineered antibodies can be further characterized for more potent and safely engineered mAb drugs
Antibody humanization has strengthened our therapeutic ability
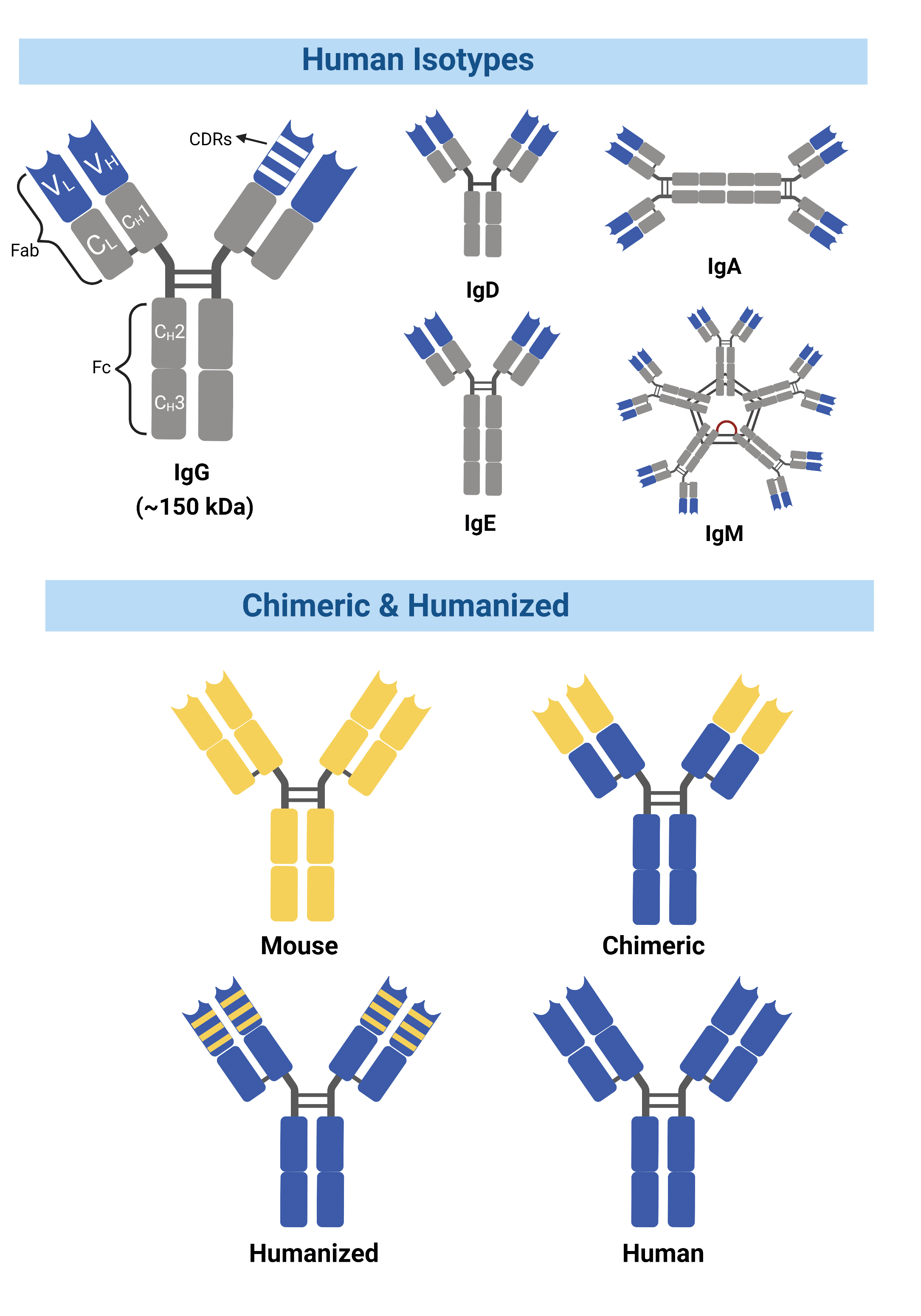
Figure 1. Chimeric antibodies contain entirely foreign variable domains, while humanized antibodies contain a smaller percentage of mouse sequences within the variable domain.
Mouse monoclonal antibodies (mAbs) are highly attractive for therapeutic applications as they allow for specific targeting while conveying effector functions and robust manufacturing procedures. However, they also pose several challenges. Notably, they have limited use as therapeutic agents because of short serum half-life, inability to trigger human effector functions and they are often recognized by the patients’ immune system as a foreign protein resulting in a human anti-mouse antibody (HAMA) response.
Antibody engineering has introduced multiple solutions to overcome these challenges. By enabling techniques that modify mouse antibodies into structures more similar to human antibodies without the loss of antigen-binding properties, it is possible to reduce the HAMA response while transforming these antibodies into suitable candidates for clinical trials. Known as chimeric, these antibodies consist of an antibody’s original antigen-binding variable domains with the constant domains from a different species (Figure 1). The first chimeric antibody, abciximab, was approved in 1994 by the FDA for inhibition of platelet aggregation. The drug was developed by combining sequences of the murine variable domain with a human constant region domain (1, 2).
Another method grafts the complementarity-determining regions (CDRs) from a mouse onto a human variable region framework, known as humanized mAbs (3) (Figure 1). The humanization of antibodies has greatly reduced immunogenicity, allowing for human effector functions to take place along with increased serum half-life in humans.
Antibody fragments and engineered variants open paths to many applications
For many applications, smaller recombinant antibody fragments and engineered variants are preferable alternatives due to their benefits such as enhanced therapeutic efficacy. Most antibody fragments are engineered by removing the constant region (Fc) due to its undesirable long serum half-life (4). The antigen-binding fragment (Fab) is the region that binds to antigens composed of one constant and one variable domain of each of the heavy and the light chain (VH, VL, CH, CL). Each variable domain contains three hypervariable loops, known as CDRs, which largely determine the antigen specificity of the antibody. A variety of antibody fragments are designed and generated using Fabs as building blocks. For example, single chain variable fragments (scFvs), a popular format comprising both VH and VL, are the smallest functional variants that retain full antigen-binding capacity. Bispecific and multi-specific antibody fragments also exist, such as diabodies and bispecific Fab2, produced by non-covalently linking different forms of Fabs or Fvs. These antibody fragments are capable of simultaneously binding two, or multiple different antigens, respectively.
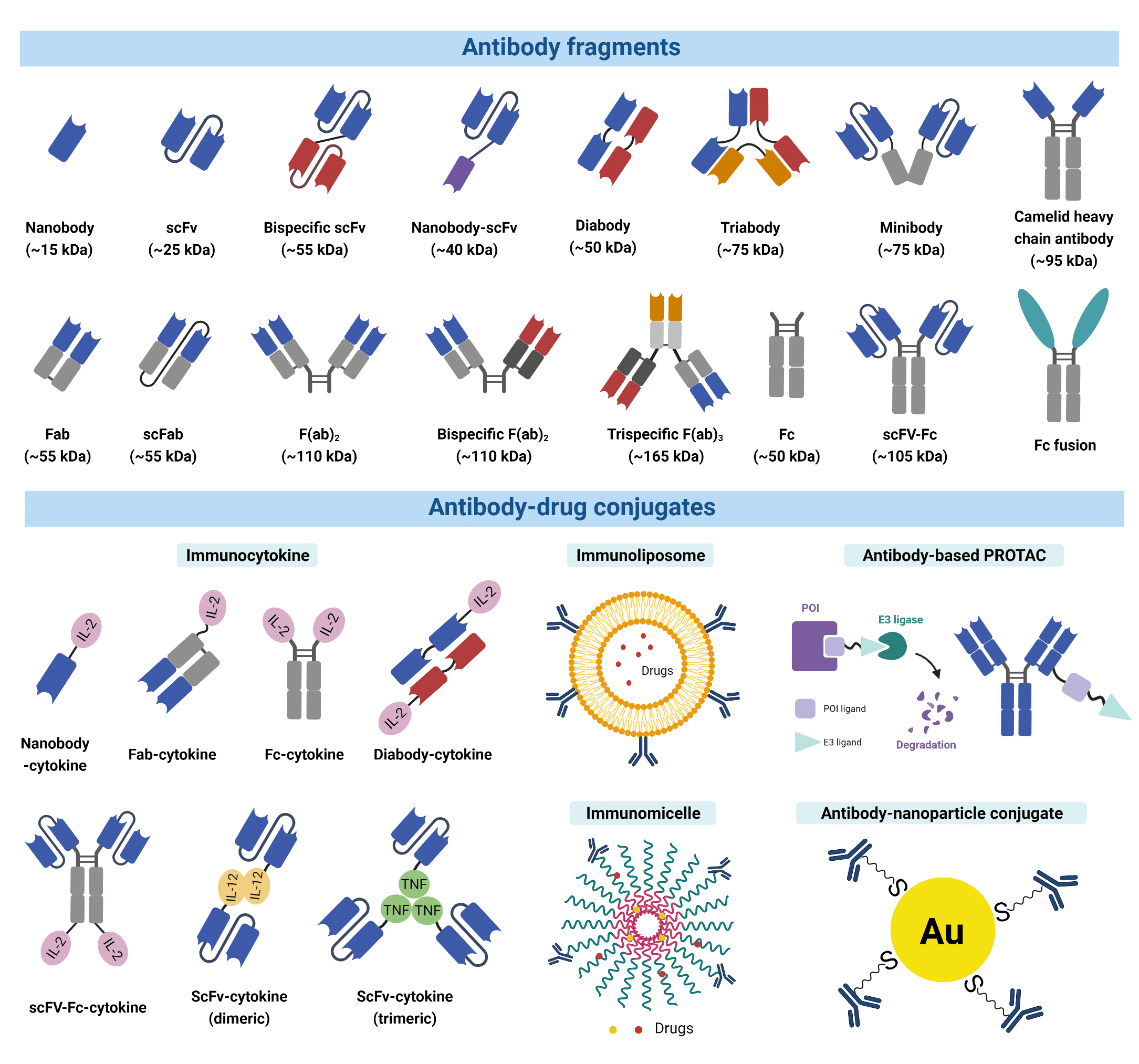
Figure 2. A wide variety of modified antibody formats and fragments are available to researchers today.
Antibody fragments and their engineered variants offer certain superiorities over a full-size antibody. For example, the smaller size of fragments permits penetration into tissues inaccessible to full-size antibodies (5,6). The lack of the Fc domain is also advantageous in immunohistochemistry and other diagnostic applications because they can greatly reduce non-specific binding to the Fc receptors, which are antibody-binding proteins found on the surface of immune cells. This non-specific binding is known to often give rise to high background signal interference. Furthermore, the fusion of multiple antibody fragments with different specificity opens up a wide range of clinical applications including directing T cells to tumour cells, dual targeting of different disease mediators, and delivering payloads to targeted sites (7).
Techniques to generate and engineer therapeutic antibodies
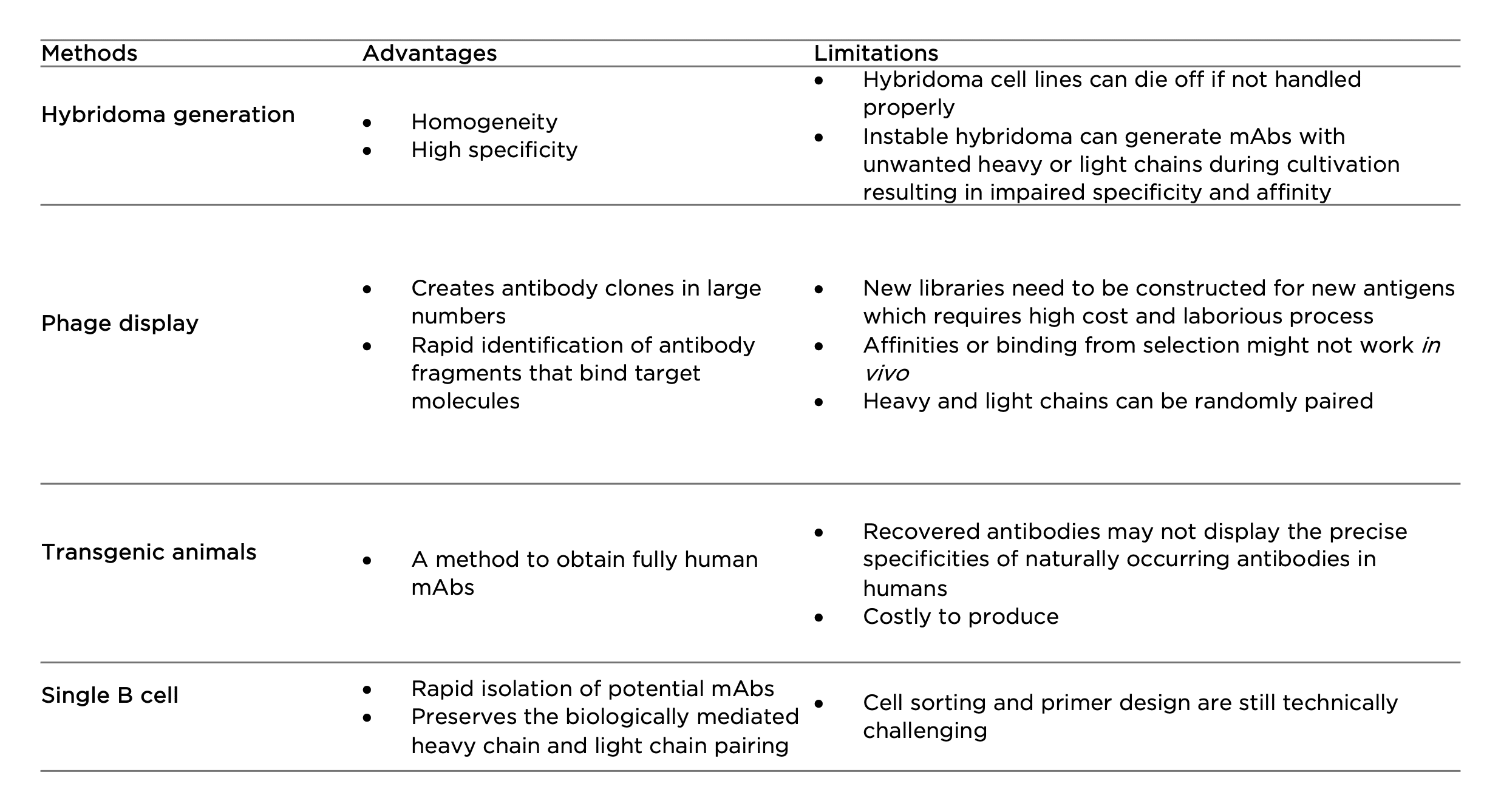
Hybridoma generation
First developed in the 1970s, hybridoma technology has remained as the primary method to produce mAbs. By fusing short-lived antibody-producing B cells with immortal myeloma cells, hybridoma cell lines are expected to provide a never-ending supply of mAbs. Hybridoma generation is widely used in antibody engineering in the initial screening and leads selection processes to further generate chimeric or humanized antibodies. The technique can also be utilized to produce bispecific antibodies by fusing two hybridomas cells to generate a single ‘quadroma’ containing different Fab or scFv modules through chemical crosslinking. However, the instability of hybridoma cell lines could cause unwanted pairing of heavy and light chains (8). The ineliminable mispairing often introduces non-functional antibody fragments, greatly hampering clinical-scale production in therapeutic antibody development.
Phage display
Phage display is the most common in vitro method used for antibody selection. It is achieved by integrating a gene sequence coding for a particular antibody into the DNA sequence of a filamentous bacteriophage. This allows fast expression of antibodies in large numbers, on the surface of the bacteriophage capsid. A library can therefore be constituted and used to detect an antigen-antibody interaction of interest. Different antibody fragments are used in phage display technology such as scFvs, Fvs, Fabs, and bispecific or bivalent antibodies. The limitation of phage display is that for each antigen a new library needs to be constructed which requires high cost and laborious process. There is also a chance that selected binders represent lower affinities in vivo. Similar to phage display, there are also yeast, bacterial, and ribosomal display technologies serving as alternative tools to address different challenges in antibody development (9).
Transgenic animals
Fully human mAbs can be generated using transgenic technology by inserting human immunoglobulin (Ig) genes into the genome of animals such as mice. Upon immunization, the animals can synthesize and produce human antibodies with low immunogenicity and high affinity for in vivo therapy. A number of human mAb drugs derived from transgenic animals have been approved by the US FDA (10). However, one disadvantage of transgenic animal systems is that they cannot precisely imitate a human immune response because of the effects of the animal’s genetic background on antigen processing and B cell regulation. As a result, the recovered antibodies may not display the precise specificities of naturally occurring antibodies in humans (11). In addition, like hybridoma technology, this technology requires substantial use of experimental animals.
Single B cell
The development of single B cells has overcome many issues raised from the above techniques. Single B cells can be isolated from peripheral blood or from lymphoid tissues against specific antigens, which is followed by mRNA extraction and direct cloning of the VH and VL region of each Ig using reverse transcription polymerase chain reaction (RT-PCR). This avoids the random pairing of heavy and light chains that often occurs in phage display (12). Following this, the genes are cloned and expressed in bacterial systems or mammalian cell lines for the immediate generation of recombinant mAbs. Once generated, the reactivity and biophysical characteristics of each antibody are determined using various assays. With the advances of next generation sequencing technology, single B cell cloning has evolved as a high-throughput solution for large-scale Ig sequence analyses and generation of antibody repertoires; this is regarded as a promising tool for developing novel mAb drugs.
Applications of de novo protein sequencing in antibody engineering
Despite the diversity of antibody engineering technologies, researchers need to be aware of the limitations of the methods they choose to use and be informed about techniques that can help them circumvent unnecessary efforts.
In general, there are two methodologies to achieve these protein engineering goals: empirical methods and rational methods. Empirical methods are based on generating large libraries and rely on screening to select the desired variants by employing hybridoma generation and phage display which usually take longer time and higher cost. Rational methods are based on the existing knowledge of antibody sequences and structures which will typically lead to generating a smaller set of variants.
Therefore, choosing rational methods over empirical methods can significantly narrow down the antibody candidate pool to minimize investment of redundant time and cost.
Antibody de novo sequencing is a good example of rational methods that uses mass spectrometry to determine the amino acid sequence of an antibody. By directly analyzing the antibody sample, this method has made direct antibody sequencing possible without the need for hybridoma cell lines or prior knowledge of the DNA sequence. The central idea of protein de novo sequencing is to determine the mass of an amino acid residue in a protein by measuring the mass difference between two fragment ions from tandem mass spectra. In such a way, the entire protein sequence can be inferred by assigning each residue along the protein backbone.
The general steps of antibody protein de novo sequencing are as follows (Figure 3):
- The antibody protein sample is digested into short peptides of overlapping sequences with multiple proteases.
- Fragmented peptides are separated using chromatographic methods.
- The mass spectra for each peptide are generated using high mass accuracy tandem mass spectrometry (MS/MS).
- Peptide de novo sequencing is conducted on the mass spectra to identify the sequence of each peptide.
- Assembly of the full antibody protein sequence is generated by data analysis with de novo sequencing software.
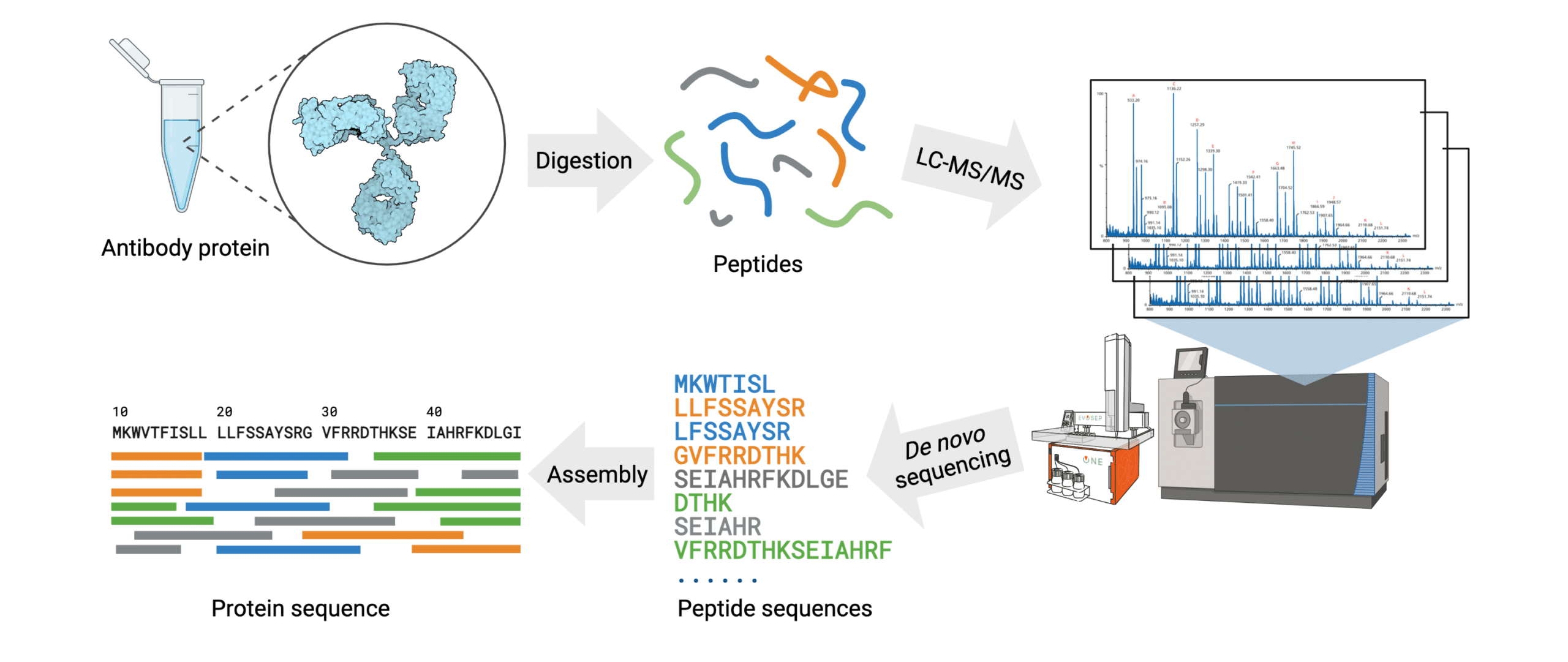
Figure 3. Workflow of the REmAb® de novo sequencing platform.
The de novo protein sequencing service, REmAb®, offered by Rapid Novor has proved its value in antibody engineering through partnerships with a myriad of projects. Listed below are some of the important applications that have been identified that help researchers more rationally develop antibody therapeutics.
(1) Antibody humanization
Typically, biopharmaceutical developers rely on hybridoma generation or phage display to develop human mAbs against human antigens of interest. However, homologous mAbs originating from other species may already be characterized and commercially available. Sequences of both commercially available or in-house generated anti-animal antibodies can be obtained with 100% accuracy and coverage using REmAb® de novo protein sequencing. With knowledge of the primary structure, different versions of the antibody may be generated to suit the research need; whether that be to engineer a chimeric version, or to tweak areas to make it a better therapeutic candidate. This application of de novo protein sequencing bypasses the generation of hybridoma cell line and phage display, which avoids the labour-intensive necessities of the empirical methods.
(2) Antibody fragments engineering
When developing therapeutic antibody fragments, or alternate variants such as bispecific antibodies or diabodies, the selection and screening process that usually relies on hybridoma and phage display can be tedious and inefficient due to the large number of choices for fragment combinations. De novo protein sequencing can be applied here to generate a series of proof-of-concept antibody fragment candidates to determine optimal target combinations. Well known and functionally validated antibodies against a variety of antigens may be de novo sequenced, which facilitates the generation of a panel of recombinant antibody fragments that engages two or more targets at once. Then In vivo and in vitro testing in primary animal and human assays can be done to determine the optimal combination of targets that are synergistic in a bispecific or multispecific format. Using well-characterized high quality antibody reagents can help safeguard the success of the early stage of a therapeutic campaign, and therefore derisking and expediting the following development (14).
(3) CAR-T Construction
Chimeric antigen receptor T cell (CAR-T) engineering is becoming quite widespread thanks to the success and hope this approach brings when no other cancer treatments have worked (15). CAR-T structures are typically composed of an ectodomain, existing in the extracellular space, a transmembrane domain, and an endodomain. Within the ectodomain, is an antigen recognition domain, typically a scFv (13). It is here where substantial research is done to determine the best target for this domain. The ideal targets should have high tumor coverage and high specificity to ensure both effectiveness and safety. However, selection of ideal targets is extremely hard to achieve in reality due to the toxic side effects toward non-tumor cells that can cause organ damage or even death (16). Thus, it is important to comprehensively assess the antibody-antigen recognition as well as the off-tumor effects. Implementing de novo protein sequencing into a CAR-T development workflow allows researchers to easily modify and create many CAR-T models by swapping the antigen recognition domain for variable region sequences found in mAbs of interest. This allows for the selection of the best scFv candidates, as well as simple engineering of these regions and detailed optimization of CAR-T models to achieve both enhanced target recognition and reduced non-tumor toxicity.
References
1. Morrison SL, Johnson MJ, Herzenberg LA, Oi VT. Chimeric human antibody molecules: mouse antigen-binding domains with human constant region domains. PNAS. 1984 Nov 1;81(21):6851–5.
2. Foster RH, Wiseman LR. Abciximab. An updated review of its use in ischaemic heart disease. Drugs. 1998 Oct;56(4):629–65.
3. Jones PT, Dear PH, Foote J, Neuberger MS, Winter G. Replacing the complementarity-determining regions in a human antibody with those from a mouse. Nature. 1986 May;321(6069):522–5.
4. Holliger P, Hudson P. Engineered antibody fragments and the rise of single domains. Nature Biotechnology. 2005;
5. K JR. Physiological barriers to delivery of monoclonal antibodies and other macromolecules in tumors. Cancer Research. 1990;50(3 Suppl):814–9.
6. Yokota T, Milenic DE, Whitlow M, Schlom J. Rapid Tumor Penetration of a Single-Chain Fv and Comparison with Other Immunoglobulin Forms. :8.
7. Fan G, Wang Z, Hao M, Li J. Bispecific antibodies and their applications. J Hematol Oncol. 2015 Dec 21;8(1):130.
8. Bradbury ARM, Trinklein ND, Thie H, Wilkinson IC, Tandon AK, Anderson S, et al. When monoclonal antibodies are not monospecific: Hybridomas frequently express additional functional variable regions. MAbs. 2018 Mar 29;10(4):539–46.
9. Ullman CG, Frigotto L, Cooley RN. In vitro methods for peptide display and their applications. Briefings in Functional Genomics. 2011 May 1;10(3):125–34.
10. Development of therapeutic antibodies for the treatment of diseases | Journal of Biomedical Science | Full Text [Internet]. [cited 2021 Feb 24]. Available from: https://jbiomedsci.biomedcentral.com/articles/10.1186/s12929-019-0592-z
11. Chiu ML, Gilliland GL. Engineering antibody therapeutics. Current Opinion in Structural Biology. 2016 Jun;38:163–73.
12. Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. Journal of Immunological Methods. 2008 Jan 1;329(1):112–24.
13. Fan C-Y, Huang S-Y, Chou M-Y, Lyu P-C. De novo protein sequencing, humanization and in vitro effects of an antihuman CD34 mouse monoclonal antibody. Biochemistry and Biophysics Reports. 2017 Mar 1;9:51–60.
14. Sen KI, Tang WH, Nayak S, Kil YJ, Bern M, Ozoglu B, et al. Automated Antibody De Novo Sequencing and Its Utility in Biopharmaceutical Discovery. J Am Soc Mass Spectrom. 2017 May;28(5):803–10.
15. Engineering CAR-T cells | Biomarker Research | Full Text [Internet]. [cited 2021 Mar 9]. Available from: https://biomarkerres.biomedcentral.com/articles/10.1186/s40364-017-0102-y
16. Wei J, Han X, Bo J, Han W. Target selection for CAR-T therapy. Journal of Hematology & Oncology. 2019 Jun 20;12(1):62.
Talk to Our Scientists.
We Have Sequenced 10,000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and developed the first recombinant polyclonal antibody diagnostics.
Talk to Our Scientists.
We Have Sequenced 9000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables timely and reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and ran the first recombinant polyclonal antibody diagnostics