March 23, 2021

*Author
#Reviewer
1Rapid Novor, Inc.
Introduction
One of the most commonly used and critical reagents found in most biomedical research laboratories is central to what is known as “The Reproducibility Crisis”. An alarming number of researchers in the life sciences have noted irreproducibility in their antibody-based experiments, fueled by insights originally unearthed by A. Bradbury and C. Glenn Begley (1–3). Through their desire to highlight the unregulated state of the antibody industry, Bradbury and Begley have helped push the need for industry reform including the need for more rigorous testing and validation.
At the root of antibody irreproducibility is the challenge of batch variability. This occurs when antibodies are sold under the same catalogue number and by the same vendor, however exhibit different specificity and/or affinity. This is often the result of cell-culturing environments and/or different producing animals. Researchers not only need to overcome the challenges associated with commercial antibody production, if they opt to produce their own, they can face a multitude of other obstacles. Notably, maintaining the high-standard procedural controls to maintain cell line viability.
The challenges associated with antibody production and supply pose a significant problem for researchers, with the worst-case scenarios preventing experimental reproducibility. In this article our team wishes to address this lack of consistency and continuity that is greatly affecting the speed and progress of scientific research from fundamental study to drug discovery and development, along with providing potential solutions.
Top 3 causes of the antibody reproducibility crisis
1. Batch-to-batch variability – a side effect of polyclonal antibody development
There are different factors contributing to the antibody reproducibility crisis. The first comes from the nature of antibody production. In human and animal bodies, antibodies are secreted by B cells in a polyclonal mixture of heterogeneous molecules that recognize and bind to different epitopes of an antigen. In the lab, polyclonal antibodies (pAbs) are produced by injecting a particular antigen into an animal that elicits an immune response. The pAbs secreted are then harvested from the animal and used in different antigen-specific experiments.
The reproducibility crisis of polyclonal-based development stems from batch-to-batch variability that commonly occurs during production. The problem arises when different host animals are injected with the same antigen but then produce pAbs that have different specificities and affinities. This means that even if one was to use a new batch of antibodies, they cannot reproduce the exact same experimental results. Even more importantly, the same animal sometimes may have different immune responses to the same antigen and also exhibit time-dependent variation known as the affinity maturation process.
2. Fragile, unstable hybridomas are endangering monoclonal antibodies
Ever since the breakthrough discovery of hybridoma technology by Köhler and Milstein in 1975, it has remained to be the primary method for scientists to produce monoclonal antibodies (mAbs). The process starts by injecting animals with specific antigen that provokes an immune response. Individual B cells producing antibodies that bind to the injected antigen are then isolated from the animal and fused with myeloma cells to produce a hybrid cell line called a hybridoma. By fusing short-lived antibody-producing B cells with immortal myeloma cells, hybridoma cells are expected to provide a never-ending supply of identical mAbs.
However, in the laboratory, hybridomas are often considered unstable and fragile, so they require high-standard procedural controls to maintain cell line viability. Although researchers spend a significant amount of time and effort on culturing hybridoma cell lines, poor growth or even cell death still occurs regularly. Under normal conditions, antibody contents can decrease continuously during the course of hybridoma cultivation (4). Many other factors can lead to failure of hybridoma survival. Long-time storage, repeated freeze-thaw cycles, improper handling, and contamination can all cause hybridoma death and permanent loss of important antibodies.
In addition, hybridoma cell lines are also known to undergo gene mutations and rearrangements over time, leading to antibody heterogeneity and batch-to-batch variability (5). A recent study of 185 clonal hybridoma cell lines identified nearly a third contain additional heavy or light chain genes, resulting in impaired affinity and specificity to the target antigen. An investigation conducted by Rapid Novor in 2019 showed similar results – among 80 research-purpose mAbs analyzed, a significant portion (14%) were shown to have a second light chain present (Figure 1). In other cases, hybridomas may even lose the chromosome containing antibody genes completely arresting the production of antibodies (6).
3. Lack of standardized validation – when antibodies mislead
Despite the complexity and uncertainty of antibody production, there is no standardized protocol or regulation for the validation of antibodies, including assessing their specificity, affinity, sequences, and applications in different assays.
Among over 2 million commercial antibodies provided by more than 300 vendors, the majority of them are polyclonal. The QC data listed on the product sheets are often obtained from previous batches and no longer valid for the ones delivered to users. With regard to mAbs, there is also no way of knowing if the current hybridoma cell line is characterized as stable and functional. This largely stems from the issue of inconsistent catalog numbers that have not accounted for new culturing environments, production animals or company merges. Lack of a proper validation process leads to the release of a large number of poorly characterized antibodies (“bad antibodies”) to users for research. It was estimated that despite there being 2 million antibodies on the market, only 12.5 – 25% are considered unique ‘core’ antibodies (7).
When it comes to selecting key antibodies, the process varies by user and laboratory. For example, some labs regularly purchase from their trusted vendors, some rely on word of mouth, and some buy a few from different vendors and then test which one works. One similarity tends to exist, many researchers will reference publications in which their antibodies were referenced or will refer to the information provided on the product sheets. However, this still does not guarantee a perfectly safe decision. Take for example the following antibodies that were used to identify therapeutically relevant clinical biomarkers. Despite all of these antibodies being regularly cited in industry publications and journals for their respective use cases, all were shown to exhibit cross-reactivity resulting in significant sums of research resources being lost (Table 1) (3).
| Target | Antibody IDs | Biomarker | Cross-reactions |
| EpoR (EPOR) | M20 and C20 | Tumor cells | HSP70 |
| ER-β (ESR2) | 12 out of 13 | Breast cancer | WDCP, POU2F1, multiple |
| HER2 (ERBB2) | 2 out of 3 | Breast cancer | HER4 |
| ERCC1 | 8F1 | Prognostic | CCT-alpha |
| CDK1 | A17 | Cancer | Cep152 |
Table 1. Some cross-reactive antibodies erroneously used to identify therapeutically relevant clinical biomarkers caused devastating personal and financial damage to science and medical progress. Adapted from reference (3).
Thus, using antibodies as biomarker tools for detection can be potentially fatal if they have not been fully verified with rigorous scrutiny. The unfortunate reality is, it’s no longer sufficient to rely solely on vendor’s quality assurance protocols or scientific publications, it is necessary to independently assess and verify candidates.
The $1.7B impact of the reproducibility crisis
It is estimated that by 2019 global spending on antibodies rose to US$3.4 billion (3), yet one study suggests US$1.7 billion may have been wasted. Within this 2008 study, 6,000 commercial antibodies were tested and only half could successfully recognize their specified targets (8). Much of the impact of the reproducibility crisis is hidden, since many researchers do not share their wasted efforts on irreproducible results. However, the impact on companies and careers can be profound. In one case, two researchers spent $500,000 and 2 years of research hours only to uncover their commercial assay for CUZD1 was actually measuring CA125 (9). Investigators around the world should validate the quality of assays and reagents even from the most reputable vendors (9).
In 2015, two articles addressing the antibody reproducibility crisis were featured on Nature and evoked extensive reaction across academia and industry. In the article Standardize antibodies used in research (8), Andrew Bradbury and Andreas Plückthun called for international collaboration to define all antibodies by their sequences and make them recombinantly to eliminate uncharacterized antibodies on the market. Indeed, less than 10% of antibodies have their sequences disclosed, for commercial reasons. Strikingly, this article had 110 co-signatories from all around the world. The second article, Blame it On the Antibodies (7), reported the results of a 2016 survey where out of 1576 researchers, 52% agreed that there was a significant reproducibility crisis and 34% did not establish any procedures for reproducibility (10).
By the end of 2015, the International Working Group on Antibody Validation (IWGAV) was founded by a group of authorities in the field of protein-binding technology with a mission to develop and formulate the best approaches for validating antibodies used in common research applications. There have also been annual international antibody validation meetings since 2014 which bring together academia and industry to establish best practices in research antibody validation and improve antibody validation for the life science community.
In 2016, IWGAV proposed a set of standard validation principles in an effort to improve antibody reproducibility, also regarded as five conceptual ‘pillars’, which include (11):
1. Immunocapture followed by mass spectrometry
2. Genetic strategies
3. Orthogonal strategies
4. Independent antibody strategies
5. Expression of tagged proteins
It was suggested in the proposal that antibody providers should validate each antibody with at least one of the above pillars in each application, and include as much additional information as reasonably possible. In response to this, many antibody manufactures are re-examining their current validation processes. Additionally, the scientific community is pushing to hold vendors accountable to defining their antibody reagents based on unique identification numbers, known as Research Resource Identifiers (RRIDs).
All these efforts will help lead to a considerable increase in research traceability and reproducibility as scientists will be provided access to information about what reagents to use and how they perform across systems, including original catalog numbers and vendor names. However, establishment of a universally reliable validation system still has a long way to go before the problem of irreproducibility can be solved.
Moving towards recombinant mAbs for a better future
Recombinant monoclonal antibodies are the most promising and straightforward solution to eliminate consistency and viability issues with antibody production and storage. Built on the development of genetic engineering, recombinant antibodies are highly consistent mAbs made by cloning the antibody DNA into an expression vector followed by producing the antibody protein in host cells such as bacteria, yeast, and mammals (Figure 2). It is an animal-free and cost-efficient method that allows for large-scale antibody production as long as the antibody sequences are known.
The 2015 article Standardize antibodies used in research has suggested a complete transition of the antibody market to recombinant antibodies, with two steps considered necessary (8). “First, the sequences should be obtained for widely used hybridoma-produced monoclonal antibodies…Second, the research community should turn to methods that directly yield recombinant binding reagents that can be sequenced and expressed easily.” Although with the current science, it is technically feasible to sequence all widely used antibodies and recombinantly produce them, it requires agreements, efforts, collaborations, and funding from all scientific sectors worldwide to achieve this transition.
Tackling reproducibility with antibody protein sequencing
The development of protein de novo sequencing has been proved to be a powerful tool for scientists looking to revive valuable but endangered antibodies. For example, N. E. Castellana and colleagues successfully reconstructed an archived antibody, lymphotoxin alpha (LT-α), a preclinical candidate for the treatment of inflammatory diseases (12). When the DNA and cell line were no longer available due to more than a decade of storage, the team used de novo sequencing technology to identify the amino acid sequence of LT-α followed by recombinant expression, with the immunoassays showing full functional activity.
Antibody protein sequencing of key antibody reagents provides researchers with a solution to rigorously test their materials prior to investing countless hours and resources into unverified products. De novo sequencing works by determining the mass difference between two fragment ions from tandem mass spectra. By first employing an enzyme digest the reagent is fragmented into smaller peptides, which can then be analysed by LC-MS/MS. From here, the spectral data can be analyzed manually, or preferably, by an automated software in order to assemble the full amino acid sequence (Figure 3).
Knowing the full amino acid sequence allows researchers to recombinantly express the antibody, enabling indefinite reproducibility with predictable biologic activity. Furthermore, antibody protein mass and sequence confirmation at scale is valuable in antibody production quality control. Compared to the hybridoma technology, recombinant antibodies have many benefits.
|
Advantages of Recombinant Antibodies |
|
| High reproducibility | The chemical synthesis process is tightly controlled to eliminate batch-to-batch variability. |
| High purity
|
The recombinant antibodies can be expressed in a chemically defined serum-free mammalian expression system. Avoiding contamination from serum components and ensuring a purity greater than 98%. |
| Decreased production time | 6 to 8 weeks v.s. 3 to 6 months. |
| Highly customizable | Isotype switching, subtype switching, species switching, reformatting to F(ab’)2, Fab can be achieved. |
| Animal welfare | The recombinant antibody manufacturing process is animal-free, thus eliminating the animal welfare concerns as well as all the costs related to animal care. |
The REmAbⓇ advantage
Rapid Novor’s proprietary de novo protein sequencing platform, REmAb®, successfully determines the primary amino acid sequences of antibodies without requiring the original hybridoma cell lines or prior knowledge of DNA sequences. By directly analyzing the antibody protein, this technology provides a solution to resurrect lost hybridomas, re-engineer proteins, and re-establish important antibodies with high consistency.
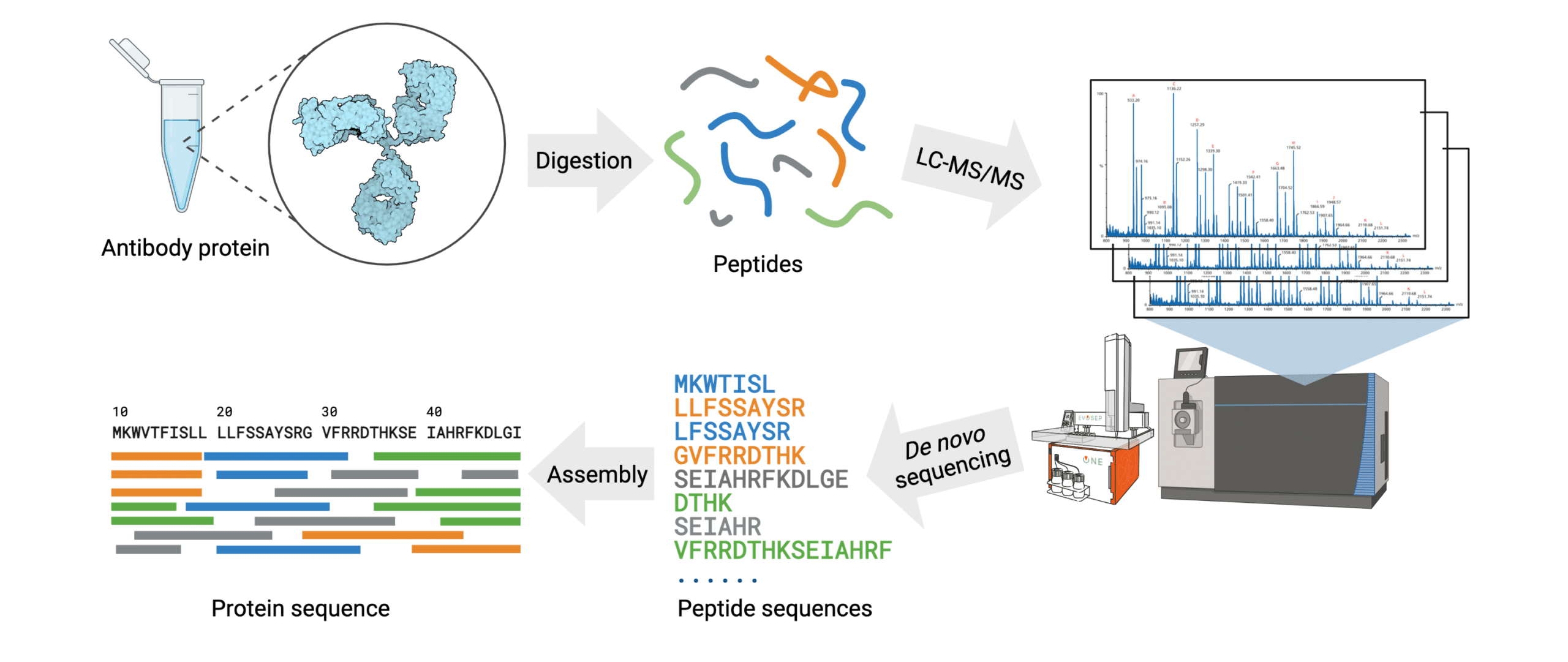
Figure 3. Workflow of the REmAb® de novo sequencing platform.
By employing their proprietary Big Data-driven machine learning algorithm, Rapid Novor assembles and generates the full antibody protein sequence within 1-2 weeks requiring only 100 μg of the protein of interest. This platform guarantees an average of at least 30X coverage for each amino acid, resulting in 100% accuracy.
Rapid Novor’s team has successfully de novo sequenced more proteins than any other organization in the world. This expertise and experience has allowed them to further push the boundaries of proteomics, leading to advancements in the likes of oligoclonal and polyclonal sequencing.
References
- Begley CG, Ellis LM. Raise standards for preclinical cancer research. Nature. 2012 Mar;483(7391):531–3.
- Bradbury ARM, Plückthun A. Getting to reproducible antibodies: the rationale for sequenced recombinant characterized reagents. Protein Engineering, Design and Selection. 2015 Oct 1;28(10):303–5.
- Voskuil JLA, Bandrowski A, Begley CG, Bradbury ARM, Chalmers AD, Gomes AV, et al. The Antibody Society’s antibody validation webinar series. mAbs. 2020 Jan 12;12(1):1794421–179441794421.
- Coco-Martin JM, Oberink JW, Brunink F, Van Der Velden-De Groot T a. M, Beuvery EC. Instability of a Hybridoma Cell Line in a Homogeneous Continuous Perfusion Culture System. Hybridoma. 1992 Oct 1;11(5):653–65.
- Castillo FJ, Mullen LJ, Grant BC, DeLeon J, Thrift JC, Chang LW, et al. Hybridoma stability. Dev Biol Stand. 1994;83:55–64.
- Bradbury ARM, Trinklein ND, Thie H, Wilkinson IC, Tandon AK, Anderson S, et al. When monoclonal antibodies are not monospecific: Hybridomas frequently express additional functional variable regions. MAbs. 2018 Mar 29;10(4):539–46.
- Baker M. Reproducibility crisis: Blame it on the antibodies. Nature News. 2015 May 21;521(7552):274.
- Bradbury A, Plückthun A. Reproducibility: Standardize antibodies used in research. Nature News. 2015 Feb 5;518(7537):27.
- Prassas I, Diamandis EP. Translational researchers beware! Unreliable commercial immunoassays (ELISAs) can jeopardize your research. Clin Chem Lab Med. 2014 Jun;52(6):765–6.
- Baker M. 1,500 scientists lift the lid on reproducibility. Nature News. 2016 May 26;533(7604):452.
- Uhlen M, Bandrowski A, Carr S, Edwards A, Ellenberg J, Lundberg E, et al. A proposal for validation of antibodies. Nature Methods. 2016 Oct;13(10):823–7.
- Castellana NE, McCutcheon K, Pham VC, Harden K, Nguyen A, Young J, et al. Resurrection of a clinical antibody: Template proteogenomic de novo proteomic sequencing and reverse engineering of an anti-lymphotoxin-α antibody. Proteomics. 2011 Feb;11(3):395–405.
Talk to Our Scientists.
We Have Sequenced 10,000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and developed the first recombinant polyclonal antibody diagnostics.
Talk to Our Scientists.
We Have Sequenced 9000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables timely and reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and ran the first recombinant polyclonal antibody diagnostics

