Written by: Genya Gorshtein, MSc
Written by: Genya Gorshtein, MSc
Published: December 12, 2024
Introduction
B cell sequencing has long been a cornerstone of antibody discovery, offering insights into the diverse repertoire of B cell receptors that drive adaptive immunity. However, this method primarily focuses on circulating B cells, potentially overlooking critical antibodies produced by plasma cells in immune reservoirs like the bone marrow.
In this article, we will compare B cell sequencing and polyclonal antibody sequencing technologies, as well as complementary approaches using proteogenomics to capture the full diversity of the antibody repertoire.
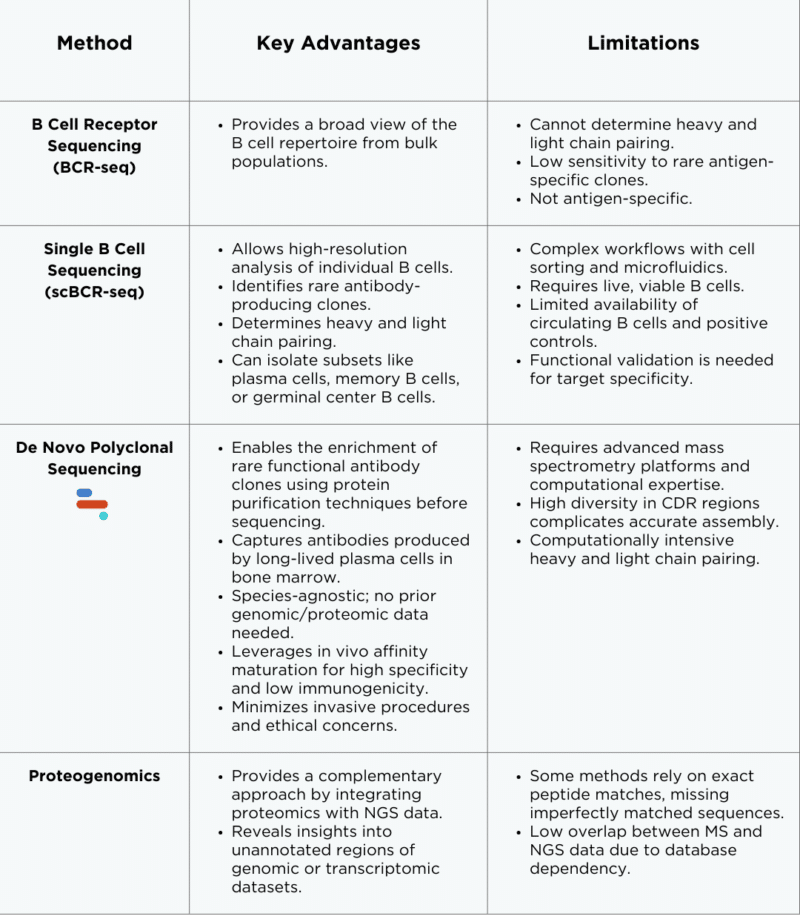
BCR and Single B Cell Sequencing for Antibody Discovery
B cell receptor sequencing (BCR-seq) and single B cell sequencing (scBCR-seq) are widely used methods for antibody discovery. BCR-seq provides a comprehensive view of the collective B cell repertoire within a sample, such as peripheral blood or lymphoid tissue, without distinguishing individual cells. In contrast, scBCR-seq isolates and analyzes individual B cells to determine their unique BCR sequences, often employing techniques like cell sorting and microfluidics for high-resolution analysis.
B Cell Receptor Repertoire Sequencing
Bulk BCR-seq is an approach that involves extracting RNA or DNA from bulk B cell populations, such as those found in peripheral blood. Using next-generation sequencing (NGS) platforms, the BCR heavy and light chain variable regions are amplified and sequenced.
Bulk BCR sequencing is well-suited for studying mature naive B cells and memory B cells. The output is a diverse repertoire of BCR sequences, often reported as a repertoire profile with frequency distributions and clonal diversity metrics.
Bulk B cell sequencing is limited by its inability to resolve the specific pairing of heavy and light chains within individual B cells, which is crucial for reconstructing functional antibodies. By examining the collective B cell repertoire in a sample, bulk sequencing provides only an aggregated view of the population, lacking the resolution needed to distinguish individual B cell receptor (BCR) sequences, or information about which clones are most relevant. Additionally, the low resolution of bulk methods can obscure the detection of rare or low-abundance antigen-specific clones, which may be crucial for antibody discovery. These limitations necessitate additional steps, such as functional validation or computational inference, to pair chains or identify specific antibodies, adding complexity and reducing efficiency.
Single B Cell Sequencing
scBCR-seq is an approach that allows for the high-resolution analysis of individual B cells. By isolating single cells through techniques such as fluorescence-activated cell sorting (FACS) or microfluidics. Unlike bulk BCR-Seq, which cannot determine the pairing of heavy and light chains within individual B cells, scBCR-Seq provides this critical information.
scBCR-seq can be broadly divided into two main approaches:
- Membrane-bound BCRs (PBMCs: Naive B cells, memory B cells; or germinal centre (GC) B cells from lymphoid tissues)
- Antibody secreting cells (ASCs) (Plasma cells or plasmablasts)
Peripheral blood mononuclear cells (PBMCs) from the peripheral blood or GC B cells can be distinguished and sorted based on cell-surface markers, such as cluster of differentiation (CD) markers, allowing for the isolation of specific B cell subsets. To assess antigen specificity, fluorescently labeled antigens in FACS are used to isolate B cells with receptors that recognize a particular target – selecting B cells with antigen specificity is a key advantage of scBCR.
In contrast, the analysis of plasma cells or ASCs typically does not involve antigen-based sorting. Instead, all ASCs are collected and screened for antigen specificity downstream. These ASCs are captured in microcapillaries and cultured in media to stimulate antibody secretion. The supernatants are then analyzed for secreted antibodies that bind the target antigen, often using antigen-coated surfaces within the microcapillary array.
Once a target-specific single B cell is identified, the genetic information for its corresponding antibodies is typically obtained by co-encapsulating the B cell with uniquely barcoded beads. These beads display oligonucleotides that have complementary sequences for the VH and VL regions, allowing the mRNA from the B cells immunoglobulin to anneal. This is followed by cell lysis, library preparation, and NGS.
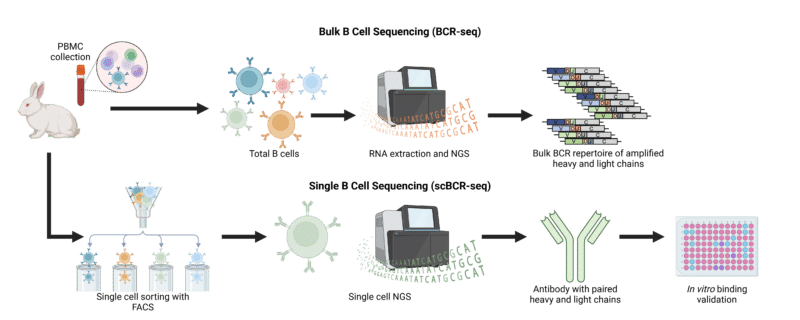
Figure 1. Workflow for bulk B cell sequencing and single B cell sequencing.
Limitations of B Cell Sequencing
B cell sequencing has become a widely used method for antibody discovery, offering valuable insights into the immune repertoire and enabling the identification of potential therapeutic candidates. However, the approach has several limitations:
- Loss of Paired Chain Information: Bulk B cell sequencing cannot determine which heavy and light chains are paired within individual B cells, which is essential for reconstructing functional antibodies.
- Lack of diversity: Circulating B cells, often used for sequencing, represent only 2-3% of the total B cell population, potentially excluding tissue-resident subsets like GC B cells or long-lived plasma cells. In the case of single cell sequencing, workflows are further limited to only approximately 10,000 cells, or approximately 0.00000001 of the available diversity at any given timepoint.
- Further loss of diversity: B cells are delicate and prone to viability loss during isolation, making it challenging to preserve their functional integrity throughout the sequencing workflow. The fragility of B cells may cause a loss of up to 75% of cells during the process.
- Primer Design Challenges: Design and availability of primers for unique animal hosts can present obstacles, especially when studying species with less well-characterized immunoglobulin gene repertoires.
- Functional Validation Required: Identified sequences must undergo functional assays to confirm antigen binding, relevant epitope, and other desired properties, adding time and complexity to the discovery pipeline.
- Lack of Positive Control: When samples contain a low number of antigen-specific cells during ASC screening, there may be no reliable positive control cells available to set fluorescence thresholds or gates for defining antigen-binding cells.
Antibody Discovery With De Novo Sequencing Serum Antibodies
De novo polyclonal sequencing allows researchers to directly analyze circulating antibodies from serum without prior knowledge of genetic information, using liquid-chromatography with mass spectrometry (LC-MS) and machine–learning algorithms.
Polyclonal antibody sequencing with mass spectrometry begins with the collection of serum, where antibodies are purified using protein A/G affinity chromatography to isolate immunoglobulins. Further antigen-specific purification is performed, often followed by protein separation using techniques such as chromatography or electrophoresis to enhance sequence resolution. The purified antibodies are digested with multiple enzymes, generating smaller peptide fragments. These fragments undergo hundreds of tandem mass spectrometry (MS/MS) runs, enabling accurate de novo sequencing to determine their amino acid sequences. Further data is gathered using top down and middle down mass-spectrometry approaches. Advanced bioinformatics tools then assemble the peptide sequences into complete heavy and light chain sequences. Finally, the heavy and light chains are paired to reconstruct the functional antibodies, providing a detailed view of the polyclonal antibody repertoire and allowing the identification of antigen-specific antibodies in circulation.
For a more in-depth description of how antibody discovery with de novo sequencing works, check out our recent publication in Nature Communications: De novo protein sequencing of antibodies for identification of neutralizing antibodies in human plasma post SARS-CoV-2 vaccination.
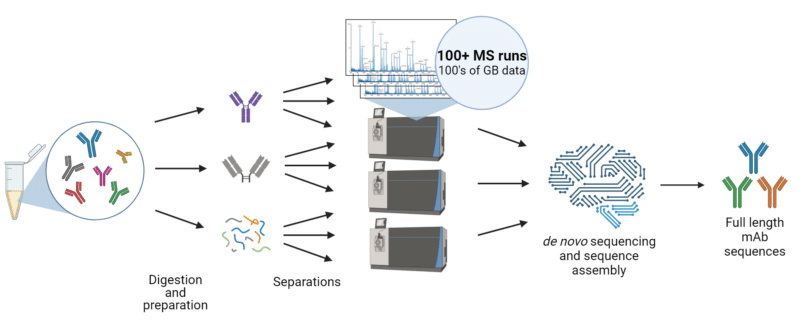
Figure 2. Workflow for polyclonal antibody sequencing for antibody discovery.
Advantages and Limitations of De Novo Polyclonal Sequencing
De novo polyclonal sequencing offers significant advantages for antibody discovery:
- Starting with Function: By starting with a population of functional antibodies present in serum, it allows researchers to enrich and purify rare clones with unique functional properties.
- Serum Advantage: Captures high-affinity antibodies produced by long-lived plasma cells in immune reservoirs like the bone marrow.
- Species Agnostic: This approach is species-agnostic, making it particularly valuable for studying unique or non-model organisms.
- High-Affinity and Low Immunogenicity: Leverages the highly evolved in vivo affinity maturation system, which generates antibodies with high specificity and low immunogenicity, unlike synthetic antibodies.
- Animal Ethics: As this method only requires serum, it minimizes the need for invasive tissue or organ collection, reducing animal use and ethical concerns.
Despite these strengths, de novo polyclonal sequencing also has limitations.
- Infrastructure and Skills: Requires specialized infrastructure, including advanced mass spectrometry platforms and computational tools, as well as highly skilled personnel to perform the analyses.
- Complex Algorithms: Algorithms are critical to assembling complementary determining regions (CDRs), antibody chains, and pairing heavy and light chains accurately, which remains a computationally challenging task.
- Technical Complexities: Polyclonal antibody populations are highly complex and require protein purification and separation techniques to deconvolute the sample and enrich for high-affinity, functional binders.
Proteogenomics: A Complimentary Approach
Proteogenomics is an antibody discovery approach that integrates mass spectrometry-based proteomics with genomic and transcriptomic data from next-generation sequencing (NGS). This method offers complementary insights into the immune and antibody repertoire, allowing comparisons between sources such as PBMCs and serum antibodies to identify repertoire convergence. However, recent attempts at proteogenomics often rely on exact peptide matches in databases, which can limit its effectiveness by excluding valuable but imperfectly matched sequences, leading to low overlap between MS/MS and NGS-derived data.
De novo sequencing addresses this limitation by bypassing the need for exact peptide matches. When integrated with NGS data, de novo sequencing can identify sequences missing from conventional NGS datasets. This capability is particularly beneficial as it directly analyzes circulating antibodies in serum, which represent the functional endpoints of the humoral immune response, offering a more comprehensive and functional perspective on the antibody repertoire.
Table 1. Advantages and disadvantages for B cell sequencing and polyclonal sequencing.
Antibody Discovery with Rapid Novor
One of the key benefits of the REpAb® polyclonal sequencing technology is the ability to sequence native antibodies directly from a purified protein or serum sample.
We recently published a research paper in Nature Communications that detailed de novo polyclonal sequencing directly from human serum, where at least 50% of the sequenced monoclonal antibodies demonstrated strong binding and functional activity.
Connect with our scientists and learn more about how REpAb® antibody discovery service can improve your antibody discovery efforts.
Talk to Our Scientists.
We Have Sequenced 10,000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and developed the first recombinant polyclonal antibody diagnostics.
Talk to Our Scientists.
We Have Sequenced 9000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables timely and reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and ran the first recombinant polyclonal antibody diagnostics