
Genya Gorshtein, MSc
Updated: October 29, 2024
(Published: July 10, 2021)
Introduction
Antibody sequencing determines the exact amino acid sequence of an antibody, providing valuable insights into its structure and function.
This process is crucial for various applications, including research and therapeutics, as it enhances research reproducibility, enables rational protein engineering, and facilitates further characterization and development.
Multiple techniques can be employed to determine and confirm the amino acid sequence of an antibody.
Sequencing an Antibody with DNA Sequencing
When the DNA sequence encoding an antibody is available, DNA sequencing technologies can be used to determine its amino acid sequence.
DNA Sequencing for Antibody Sequencing
This process typically involves amplifying the specific gene that encodes the antibody of interest using polymerase chain reaction (PCR) to generate sufficient quantities for analysis. Once amplified, next-generation sequencing (NGS) or Sanger sequencing can be employed to read the nucleotide sequence accurately. After obtaining the DNA sequence, the sequence can be translated into the corresponding amino acid sequence of the antibody.
Utilizing DNA sequencing techniques is not always feasible. Unless the hybridoma cell line or recombinant cell line is healthy and viable, retrieving the DNA sequence can be difficult. In fact, the DNA sequence is often unavailable or provides inadequate information to determine the antibody sequence.
Several factors contribute to the unavailability of the DNA sequence:
- Proprietary Information: Many commercial antibody producers consider the DNA sequence proprietary, limiting access to their sequences.
- Hybridoma Cells: In some cases, hybridoma cells used to produce monoclonal antibodies may be dead or unavailable, making it impossible to retrieve the DNA sequence.
- Natural Antibodies: Antibodies derived from natural sources, such as serum or tissues, may not have an associated DNA sequence available for analysis.
- Complex and Engineered Antibody Formats: Antibody formats, such as bispecific antibodies or antibody-drug conjugates, may have complicated structures that make it challenging to trace back to a DNA sequence.
There are ongoing efforts to create antibody databases that store validated antibody sequences of reagent mAbs. Such databases include: IMGT/mAb-DB, abYsis, or The ABCD (for AntiBodies Chemically Defined) database.
DNA Sequencing is Sometimes Not Enough for Antibody Sequencing
Even if the DNA sequence of hybridoma cells or recombinant cells is available, the DNA sequencing alone is not sufficient for antibody sequencing. Several factors related to protein structure and expression are not captured by DNA sequencing alone, as outlined below:
- Post-Translational Modifications: Antibodies undergo various post-translational modifications (PTMs) such as glycosylation, phosphorylation, and methylation. These modifications can significantly influence the antibody’s structure, function, and stability. DNA sequencing does not provide information about these critical modifications.
- Alternative Splicing: Antibodies can be generated from the same gene through alternative splicing, resulting in different isoforms with distinct properties. DNA sequencing will only reveal the sequences present in the genome, not the functional protein variants produced.
- Sequence Variants: Recombinant expression systems or hybridomas can sometimes produce sequence variants that differ slightly from the intended antibody sequence due to errors during transcription or translation. These variants may not be reflected in the original DNA sequence, but they can influence the antibody’s binding properties or biological function.
- Hybridoma Instability: Hybridomas can exhibit genetic instability over time, leading to spontaneous mutations or chromosomal aberrations that alter the antibody sequence. This instability may result in the production of non-functional or modified antibodies, often characterized by additional light chains or oligoclonal-like expression. These variations can complicate DNA sequencing efforts and hinder accurate antibody characterization.
Protein Sequencing for Obtaining Antibody Sequences
The most effective method for sequencing antibodies is mass spectrometry-based protein sequencing. LC-MS techniques enable researchers to directly determine the amino acid sequences of proteins from biological samples, often without requiring prior genetic information.
In some cases, mass spectrometry-based sequencing can cross-reference MS data with antibody databases to identify the antibody sample. However, due to proprietary ownership and the lack of fully sequenced genomes, many antibody sequences are not publicly available. While researchers can use these databases to find sequences of closely related antibodies and rely on bioinformatics tools to make predictions about the antibody of interest, this approach is not ideal for conducting confident and rational research.
De Novo Antibody Sequencing
If the antibody sequence is not available in any antibody databases, de novo antibody sequencing using mass spectrometry is the only solution for obtaining the amino acid sequence.
This approach employs liquid chromatography coupled with mass spectrometry to derive the antibody sequence solely from the resulting mass spectra data, without relying on any database or prior sequence knowledge. As the most accurate and unbiased method for obtaining antibody sequences, de novo sequencing can also account for post-translational modifications and sequence variants.
How Do You De Novo Sequence an Antibody?
The workflow for de novo antibody sequencing generally follows these steps:
- Sample Preparation: The purity of the sample is assessed using SDS-PAGE. This determines whether additional purification methods are needed to improve sample quality for sequencing.
- Enzymatic Digestion: Multiple proteases are selected and used to enzymatically digest the antibody into short peptides. Utilizing different enzymes generates peptides of varying sizes and overlaps, ensuring adequate sequence coverage in subsequent steps.
- Liquid Chromatography with Tandem Mass Spectrometry (LC-MS/MS): High-performance liquid chromatography (HPLC) is used to separate the peptides, which are then analyzed using a mass spectrometer to determine the mass to charge ratio. Care is taken in selecting the fragmentation method for MS2 and the analysis window to ensure optimal coverage.
- Peptide De Novo Sequencing: Each mass spectrum is interpreted to determine the sequence of each peptide. While software is available for this task, the latest algorithms provide faster results with less ambiguity. Expert human interpretation is often necessary for post-software analysis.
- Sequence Assembly: The full-length protein sequence is constructed from the peptide sequences, aiming for maximum overlap. Expert human interpretation may still be required after software analysis to ensure accuracy.
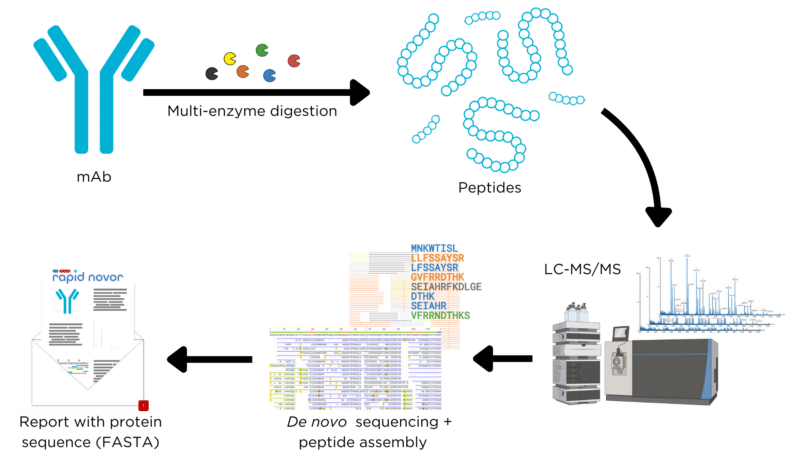
Figure 1. Workflow for LC-MS-based protein sequencing.
Monoclonal vs. Polyclonal Sequencing
Protein sequencing for antibody identification is feasible for both monoclonal and polyclonal antibodies. Polyclonal antibody sequencing allows for the conversion of polyclonal antibody reagents into characterized monoclonal antibody cocktails that replicate the original polyclonal mixture. This approach reduces batch-to-batch variability while preserving the advantages of polyclonal antibodies for use in in vitro assays.
However, polyclonal antibody sequencing involves greater complexities due to the diverse nature of the sample, which can contain several to hundreds of different heavy and light chains that must be accurately paired to form functional antibodies.
The sequencing process for polyclonal antibodies resembles that of monoclonal antibodies but requires more intricate chromatography separation and purification techniques (Figure 3). Typically, it involves 100 to 500 mass spectrometer runs, generating hundreds of gigabytes of data. This information is processed using advanced machine learning algorithms to accurately assemble hundreds of peptide sequences into individual, full-length antibody sequences with paired heavy and light chains.
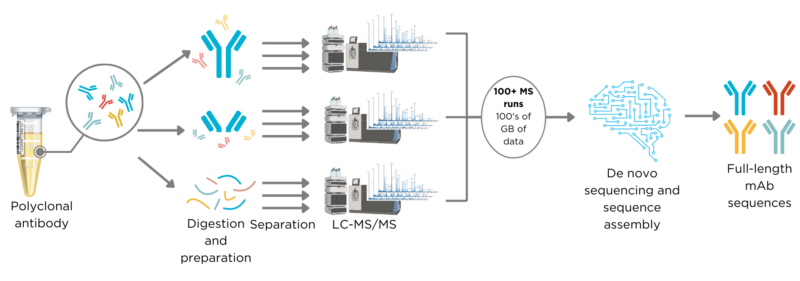
Figure 2. Workflow for de novo polyclonal antibody sequencing using REpAb®.
Rapid Novor Antibody Sequencing Services
De novo antibody sequencing is no easy task, as it requires interdisciplinary expertise in protein biochemistry, proteomics, mass spectrometry, and machine learning algorithms to obtain the most accurate protein sequence.
Rapid Novor is the world leader in antibody and protein sequencing, built on 25+ years of research, proteomics, and bioinformatics research.
- REmAb® monoclonal antibody sequencing requires only 50μg of sample to derive the complete amino acid sequence with 100% accuracy and coverage in 2 weeks or less.
- REpAb® polyclonal antibody sequencing requires only 1 mg polyclonal antibody and returns full length and paired heavy and light chains.
Rapid Novor also offers a comprehensive suite of mass spectrometry-based characterization services for antibodies including: sequence variant analysis, PTM analysis, glycan analysis, peptide mapping sequence confirmation, and more.
Contact us to learn more.
Talk to Our Scientists.
We Have Sequenced 10,000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and developed the first recombinant polyclonal antibody diagnostics.
Talk to Our Scientists.
We Have Sequenced 9000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables timely and reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and ran the first recombinant polyclonal antibody diagnostics

