Zac McDonald1, Signy Chow2,4, Kathleen Gorospe1, Xin Xu1, Paul Taylor1, Qixin Liu1, Zhihua Li2, Ziwei Han5, Trevor J. Pugh2,3, Suzanne Trudel2, Bin Ma5
1Rapid Novor Inc., Kitchener, ON, Canada
2Princess Margaret Cancer Centre, University Health Network, Toronto, ON, Canada
3Ontario Institute for Cancer Research, Toronto, ON, Canada
4Sunnybrook Health Sciences Centre, Toronto, ON, Canada
5University of Waterloo, ON, Canada
Abstract
The DNA sequences of antibodies are highly diverse due to the V-(D)-J recombination and hypersomatic mutations. As such, relying on homology-based searches to sequence novel antibodies can introduce bias to sequences obtained from proteomics approaches. De novo sequencing of antibody proteins directly from MS data is an improvement over the conventional database approach, but recent de novo sequencing studies are based on very few antibodies that may not be representative of samples. Here, we conduct the first large scale test of the REmAb® sequencing platform by comparing its protein and DNA sequencing results of 24 myeloma cell lines.
Key Takeaways
- REmAb® de novo protein sequencing service is capable of fully sequencing antibodies comparably to DNA sequencing
- REmAb® could fill in the gaps of sequences accurately even in cases where DNA sequencing could not
Introduction
The DNA sequences of antibodies are highly diverse due to the V-(D)-J recombination and hypersomatic mutations. As such, a new antibody of interest is unlikely to appear in any existing sequence database. Consequently, the database search approach commonly used in proteomics does not work for antibodies. De novo sequencing of antibody proteins directly from MS data is an improvement over the conventional database approach. However, current published de novo sequencing methods are based on data from few, selected antibodies that may not be representative of samples. Here, we conduct the first large scale test of the REmAb® sequencing platform by comparing its protein and DNA sequencing results of 24 myeloma cell lines.
Materials & Methods
The cells were grown in Gibco™ IMDM medium supplemented with 1% FCS. Supernatants were collected after 72 h in culture. After removal of BSA, samples were digested with Trypsin, LysC, Chymotrypsin, Pepsin, and AspN (Promega, WI, US). MS data were collected with Orbitrap Fusion. Protein sequences were assembled with the REmAb® sequencing platform. The Ile and Leu were determined with the WILD® method. The DNA sequences were generated using a novel hybrid capture approach using custom probes designed to target all annotated alleles of the V, J, and C regions of the IMGT database. The tBLASTn program was used to translate the DNA reads and align them with the protein sequence to check the correctness of protein sequencing results.
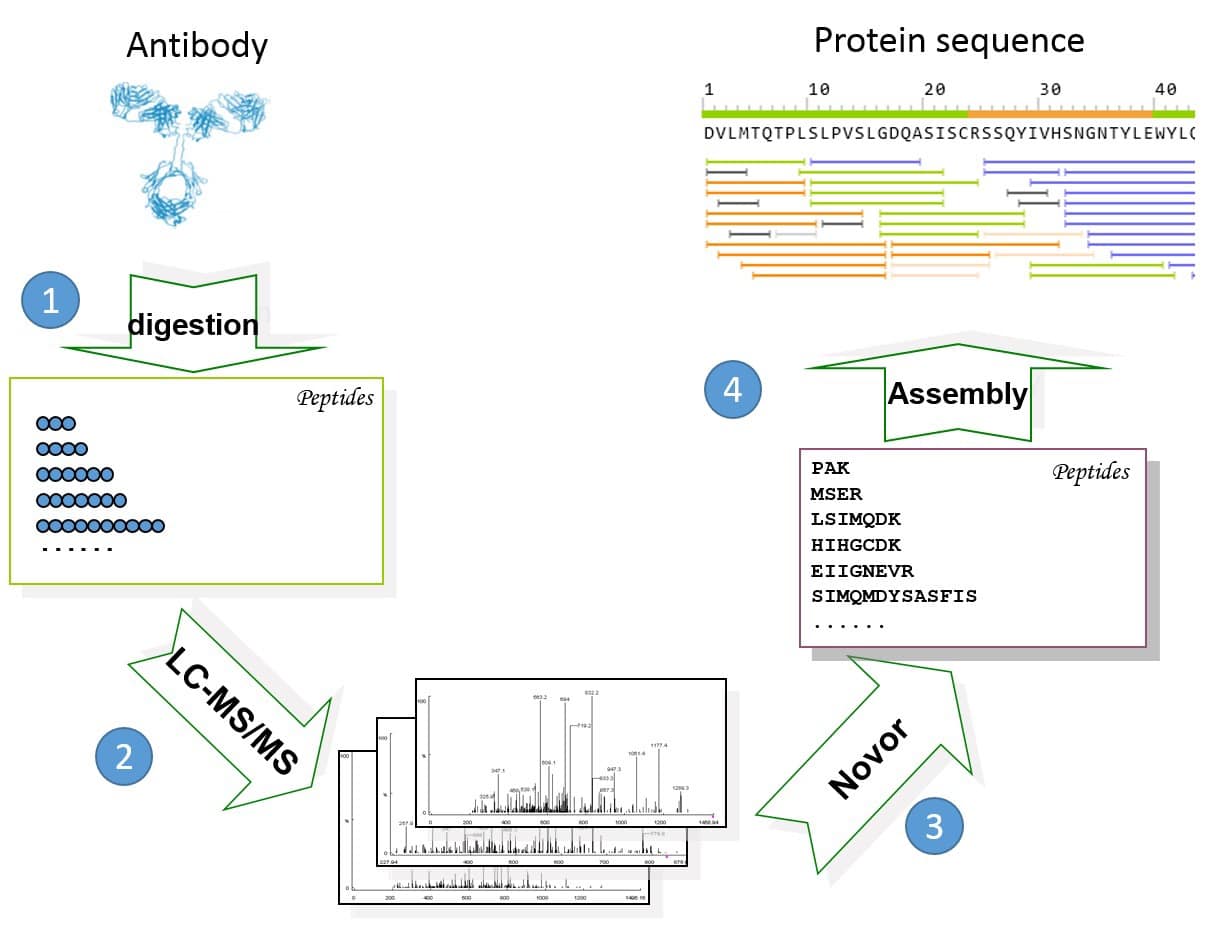
Figure 1. General workflow of REmAb® includes (1) multiple enzyme digests, (2) mass spec, (3) de novo peptide sequencing, and (4) sequence assembly.
Results
Twenty-four Myeloma cell lines have been processed thus far. Two cell lines (KMS-12BM and UTMC2) failed to express antibodies (verified by intracellular flow cytometry and MS independently). Two (AMO-1 and H929) produced a very low amount of antibodies requiring additional enrichment in future for sequencing, leaving 20 cell lines that produced sufficient antibodies for MS-based sequencing using our standard protocol. Four cell lines (EJM, LP1, OCI-My1, OCI-My6) express both heavy and light chains, whereas the other 16 express only light chains.
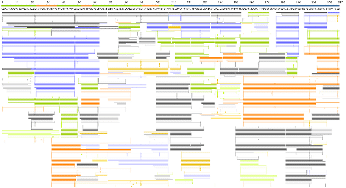
Figure 2. Screenshot of REmAb® software showing the coverage of the protein sequencing result of cell line OCI-My5. The top sequence is obtained with REmAb®. Each colored bar below the sequence indicates a unique peptide-spectrum match (PSM) covering the area. Different colors indicate different enzymes used for proteolysis. The actual coverage depth is greater than shown by the screenshot as the figure is cropped.
For each of the 20 protein samples, 5 LC-MS/MS runs were performed on an Orbitrap Fusion instrument. On average each LC-MS run produced between 5,000-10,000 MS/MS spectra, including HCD, ETD and EThcD spectra. The data is of high quality, with mass error of no more than 3 ppm for most spectra. All of the expressed heavy and light chains were sequenced with high confidence on the REmAb® sequencing platform. Figure 1 shows the protein sequence of cell line OCI-My5 and its MS/MS spectra coverage. Each amino acid is supported by tens of unique peptide-spectrum matches (PSM). A PSM is unique if it has a unique combination of its sequence, PTM, charge state, and fragmentation parameter. The coverage of other cell lines is similar.

Figure 3. Screenshot of the alignment between the protein sequence (top line) and the translation of the DNA sequencing reads (bottom lines) for OCI-My5. The number after each read indicates the number of identical reads aligning to the same location. The actual coverage depth is greater than the screenshot is shown as the figure is cropped.
Conclusions
By using the right MS experiments and software tools, antibody proteins can be de novo sequenced routinely. The isobaric Ile and Leu can also be distinguished with w-ions from EThcD spectra.
This case study was adapted, with permission, from McDonald, Z., Chow, S., Gorospe, K., Xu, X., Taylor, P., Liu, Q., Li, Z., Han, Z., Pugh, T., Trudel, S., & Ma., B. (2019). A Large-Scale Comparison of MS-based Antibody De Novo Protein Sequencing and Targeted DNA Sequencing. ASMS 2019 Atlanta, WP 046.
Talk to Our Scientists.
We Have Sequenced 10,000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and developed the first recombinant polyclonal antibody diagnostics.
Talk to Our Scientists.
We Have Sequenced 9000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables timely and reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and ran the first recombinant polyclonal antibody diagnostics

