Dr. Thierry Le Bihan joined Rapid Novor as a Principal Scientist in 2018, tasked with leading a multidisciplinary team to develop a polyclonal antibody sequencing platform. Thierry brought extensive proteomics and mass spectrometry expertise from his previous roles as Director of Proteomics at The Campbell Family Institute for Breast Cancer Research at The Princess Margaret Cancer Centre and as an Associate Professor at the University of Edinburgh.
With his out-of-the-box thinking approach, Thierry has driven innovation in the antibody discovery space and led his team through several breakthroughs, including sequencing functional antibodies from human blood.
I sat down with Thierry to ask about his approach to polyclonal sequencing, challenges along the way, and the future of antibody discovery.
What was your previous research experience and area of expertise prior to joining Rapid Novor?
Before joining Rapid Novor, I was an Associate Professor at the University of Edinburgh. My research there primarily focused on the proteomic analysis of the algae Ostreococcus, one of the most primitive organisms next to yeast. This work was intellectually stimulating and involved both basic biological research and collaboration with astrobiologists to explore the possibilities of life on other planets. Despite the exciting nature of this research, I found it increasingly difficult to see a clear path for practical applications, and it lacked the element of risk that drives innovation.
I soon found Rapid Novor. I spoke with the co-founders, Dr. Bin Ma, Mingjie Xie, and Qixin Liu. and it was obvious to me that they had a very clear vision for the future of Rapid Novor, with real-world applications that could really make a difference.
Dr. Bin Ma, the Chief Scientist, in the early-day discussions mentioned polyclonal antibody sequencing – something nearly unheard of over 6 years ago. There were two papers at the time that attempted to de novo sequence polyclonal antibodies. We asked ourselves ”What if we could do it the best?” Surely we had the bioinformatics, and de novo antibody sequencing expertise to tackle this.
We slowly, over the course of 6 years have established a successful and validated de novo polyclonal sequencing platform. We are a team of 4 laboratory technicians and 4 senior scientists – who are the pillars of the polyclonal sequencing team, and their dedication has been instrumental in our success.
What were some of the biggest challenges you faced in de novo sequencing polyclonal antibodies, and how did you address them?
Our biggest challenge was in the early stages of platform development. How do you establish a first-of-its-kind polyclonal sequencing workflow?
When you establish a platform, you start with basic samples and experiment with different types of conditions and preparation techniques. You run the samples on the mass spectrometer, analyze the data, and that data serves as feedback to inform your sample preparation procedures. You’re always chasing that feedback.
We could generate and process samples, but there was no satisfying way to analyze the data, and thus, no feedback. Are you going in the right direction? You don’t know, and you have to pause until you can answer this question. That’s what we did—we would pause one effort and work on something else, and at some point, come back to the original question with a solution. We developed an algorithm, based on Novor, our monoclonal antibody sequencing platform to analyze and de novo sequence polyclonal antibodies based on the mass spectra. Over time, we tackled difficulties such as assembling highly variable CDR sequences and pairing heavy and light chains with the combination of bioinformatics and biochemistry techniques.
We are experiencing the same thing with NovorIg now, a platform to monitor the immune repertoire over time. Only recently have we begun finding solutions and strategies to generate feedback on this platform, analyze the data and establish a validated system.
When you buy a tool antibody, it comes with a reagent sheet and instructions for its use in specific assays. However, with polyclonal antibody samples, whether purified proteins or serum, there are no instructions. It’s up to us to understand how to work best with each sample, extract the most information, and return valuable insights to our customers.

Can you discuss a recent project or discovery in your lab that you are excited about?
The most exciting projects are the ones with the potential to make a significant impact. Much research is focused on infectious diseases such as Lyme disease, measles, malaria, SARS-CoV-2, and many others. The ability to discover antibodies generated from immune responses to these diseases and create immunotherapies and other therapeutic options is incredibly motivating.
In the early days of platform development, small victories were the most thrilling. I remember the first polyclonal antibody we sequenced—I couldn’t sleep that night. We sequenced mAbs from a polyclonal mixture, recombinantly expressed them, and tested them with ELISA. To our excitement, the mAbs bound to the antigen with the same affinity or better than the original pAb mixture. This was a huge milestone for us at the time. Other exciting discoveries have followed incrementally, such as the first antibody discovery against a full protein versus a small peptide, or isolating different isotypes from alpaca immunoserum.
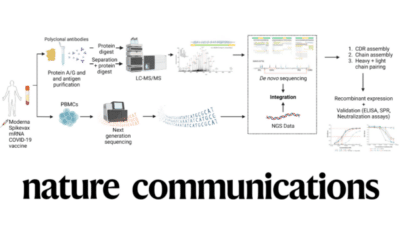
Recently, we completed an antibody discovery project in humans by sequencing functional and neutralizing antibodies from patients vaccinated against Covid-19. This project is particularly exciting for us since the human antibody repertoire is incredibly diverse.
How do you foster innovation and collaboration within your team?
I believe the key is to let people pursue what they enjoy and excel at. Encouraging the polyclonal sequencing team to foster their own ideas and experiment with new methods and techniques is crucial. Innovation often stems from creativity and nurturing those crazy and out-of-the-box ideas. If one of the laboratory technicians has a “crazy” idea, let’s try it. Sometimes, that crazy idea results in a novel purification technique or inspires other ideas that solve technological problems and expand our capabilities. Our team isn’t afraid to try something new.

What do you envision for the future of antibody discovery? What major breakthroughs do you foresee on the horizon?
My dream for the future of antibody discovery is to establish a personalized proteomics approach, similar to personalized genomics tests, to understand individual immune repertoires. I envision analyzing an individual’s IgGs to uncover the relationships between environment, disease, background, and hereditary factors on immune responses and IgG repertoires.
We know that the human antibody repertoire is incredibly diverse, but how unique is an individual’s immune repertoire compared to others? This information could be critical for addressing major questions related to autoimmune diseases, cancer, and risk assessment for infectious disease. I hope that in the next 10 to 15 years, our polyclonal antibody sequencing and bioinformatics technology will mature enough to provide these answers.
The polyclonal sequencing and discovery team has worked with several different species, what’s your dream species to work with and why?
I believe everyone at Rapid Novor, and maybe some of our clients, knows that my dream species to work with is sharks. As one of the oldest living species with adaptive immune systems, sharks generate unique IgNAR antibodies with heavy chains only. This aligns with my curiosity to understand the relationships between different species and their unique immune responses and antibody repertoires.
We can explore questions such as the relationship between IgGs and species, or the relationship between antibody repertoires and geographical locations of humans or animals. There is a lot to learn about different species, their evolution, and their unique and diverse repertoires, and what we can discover and engineer.

Published July 31, 2024
Learn More About Our Antibody Discovery Technology
Pioneered by Rapid Novor, polyclonal antibody sequencing with REpAb® is an emerging antibody discovery service that derives full-length mAb protein sequences directly from complex polyclonal populations. REpAb® seamlessly integrates mass spectrometry-based proteomics and big data learning to identify and characterize the biologically relevant, circulating antibodies from their native source.
Read our recently published paper in Nature Communications on Sequencing Neutralizing Antibodies Directly from Human Serum.
Le Bihan, T., Nunez de Villavicencio Diaz, T., Reitzel, C. et al. De novo protein sequencing of antibodies for identification of neutralizing antibodies in human plasma post SARS-CoV-2 vaccination. Nat Commun 15, 8790 (2024).
Talk to Our Scientists.
We Have Sequenced 10,000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and developed the first recombinant polyclonal antibody diagnostics.
Talk to Our Scientists.
We Have Sequenced 9000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables timely and reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and ran the first recombinant polyclonal antibody diagnostics