
Published: August 8, 2024
Introduction
Sequence variants are unwanted impurities in antibody and biotherapeutic products that can significantly impact their physicochemical and biological properties. These impurities can affect the drugs efficacy and safety, posing potential risks to patients. As such, regulatory agencies have stringent expectations regarding the quality and extent of data and evidence related to sequence variants and their associated risks.
Detecting sequence variants early in drug development is challenging due to their low abundance and the complexity of identifying these subtle alterations. This difficulty underscores the importance of employing advanced analytical techniques to ensure the purity and consistency of biotherapeutics throughout the development process.
What are Protein Sequence Variants
Protein sequence variants are the result of unintended amino acid substitutions in biologic drugs, such as monoclonal antibodies. There are two main types of sequence variants (figure 1):
- Mutations: Arise from DNA-level errors and nucleotide mutations during DNA replication processes. Typically caused by error-prone DNA repair mechanisms, frame shifts, deletions, and rearrangements.
- Misincorporations: Arise from incorrect mRNA incorporations during transcription (DNA → mRNA) or incorrect amino acid incorporations during translation (mRNA → protein). These misincorporations are the result of non-optimized codon usage, tRNA mischarging, or codon-anticodon mispairing.
Naturally occurring sequence variants in humans occur in very low levels, at approximately 0.01% sequence variants in a given protein population. Generally, these naturally occurring sequence variants are not cause for concern. However, the occurrence of sequence variants can increase significantly in recombinant protein expression systems, especially in early production stages.
In well-optimized recombinant production systems, the levels of sequence variants are at approximately 0.08%. In recombinant expression systems without fully optimized cell culture and bioreactor conditions, the rate of sequence variants can be as high as 0.1% to 50%. In recombinant expression systems, the main cause of misincorporations is due to unbalanced cell culture conditions where nutrients and amino acid availability causes substitutions. Often, amino acid supplementation and cell culture optimization can mitigate these substitutions.
Sequence variants are a cause for concern even at low levels and the occurrence can significantly increase in early production stages. Removal of sequence variants during purification stages of antibody development is extremely difficult. As such, it is more feasible to detect the presence of sequence variants and correct cell culture conditions early. Careful optimization of cell culture and bioreactor conditions minimizes risk of sequence variants, in addition to conducting sequence variant analysis during early and late stages of product and process development.
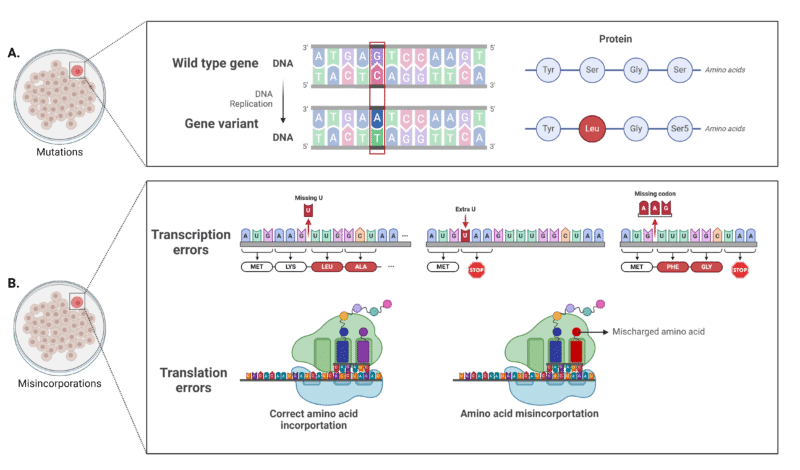
Figure 1. Types of sequence variants. A. Mutational sequence variants that arise from genetic mutations during DNA replication. B. Misincorporations due to transcription or translation errors.
Impact of Sequence Variants on Monoclonal Antibodies
Sequence variants can potentially lead to protein misfolding, changes in biological function, reduced efficacy, aggregation, and unknown immunogenic effects. These variants are highly heterogeneous due to the possibility of any amino acid substitution in the protein sequence. This heterogeneity complicates the characterization of their impact on the efficacy and safety of biologics, as the functional consequences can vary greatly. Characterizing the effects of translational errors and sequence variants in vivo does not directly translate to biotherapeutics due to various factors, including bioavailability, dosage, administration route, and the drug’s clearance mechanisms.
Some sequence variants have predictable functional effects. For example, a sequence variant in a complementarity-determining region (CDR) is likely to impact antigen binding affinity, while a variant in the Fc domain is likely to affect binding to Fc receptors. The effects of sequence variants must be analyzed on a case-by-case basis.
A previous study showed that a sequence variant of anti-HER2 IgG1, with a post-translational isomerization of Asp102 in the HCDR3 region, reduced the drug’s potency by 70%. In a different study, an amino acid substitution in the light chain from Asn35 to Ser resulted in only a minor decrease in antigen binding affinity.
The effects of safety and immunogenicity of sequence variants remains unanswered. It is difficult to predict immunogenicity for biologics, as immunogenic reactions can arise from any portion of the biotherapeutic, making it challenging to assign them to a specific sequence variant. A previous study described that the isomerization of aspartic acid to isoaspartic acid in a tyrosine-related protein triggered a strong immune reaction. This highlights the importance of ongoing research and vigilance in the monitoring of biotherapeutic products to ensure patient safety.
Challenges in Sequence Variant Analysis and Detection
One of the primary challenges in identifying sequence variants is their low abundance, which makes them difficult to detect using conventional analytical methods. Early in drug development, these variants may be present at levels that are not easily discernible, requiring highly sensitive and specific techniques to identify and quantify them.
The sheer number of possible sequence variants adds another layer of complexity. Each amino acid substitution generates a new variant, leading to a vast number of possible combinations that need to be screened. This high variability necessitates comprehensive and detailed analysis to ensure that all potential variants are identified and characterized.
Techniques such as mass spectrometry, next-generation sequencing, and advanced chromatography play a critical role in the detection and characterization of sequence variants.
Sequence Variant Analysis by Mass Spectrometry
Tandem mass spectrometry is one of the most commonly used tools for detecting sequence variants at the peptide level (Figure 2). The experimental setup and conditions are similar to peptide mapping, but the key difference lies in the analysis. Unlike standard peptide mapping, this approach requires searching for sequence variants that deviate from the expected sequence. Only a few specialized softwares are capable of identifying unexpected mass shifts as modifications or sequence variants. Due to the high number of possible substitutions and sequence variants, there is a high rate of false positives for sequence variant identification. For example, in one study, 7 possible sequence variants were reported by the analysis software, but manual assessment revealed that only 1 variant turned out to be correct.
While mass spectrometry is a highly sensitive technique, sequence variant analysis takes a trained eye to ensure accurate analysis and variant identification. Liquid chromatography is often used to separate small amounts of variants, but this process demands big columns and high sample volumes.
For more information on using LC-MS for sequence variant analysis and antibody characterization, watch our on-demand webinar: Ask Our Experts: Navigating Antibody Characterization through LC-MS.
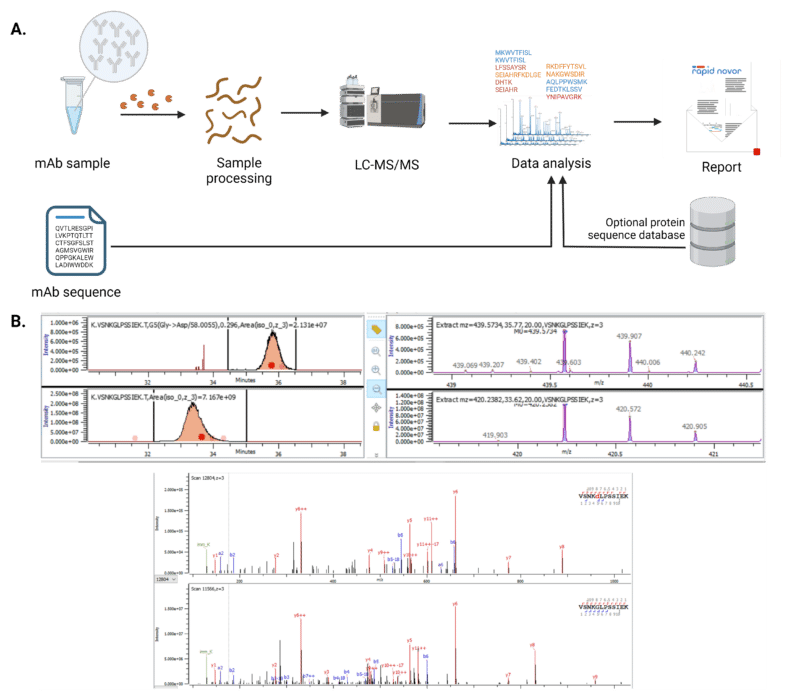
Figure 2. Mass spectrometry is a common method for sequence variant analysis at the peptide level.
A. Workflow for sequence variant analysis by mass spectrometry. B. Mass spectrometry output
Sequence Variant Analysis by Next Generation Sequencing
Next generation sequencing (NGS) technology is often used to identify sequence variants at the DNA or RNA level. NGS enables the simultaneous sequencing of millions of DNA or RNA fragments, allowing for the comprehensive analysis of complex mixtures of sequences. This high-throughput capability is particularly beneficial in identifying sequence variants that may be present at very low levels within a sample. This includes information on single nucleotide polymorphisms (SNPs), insertions, deletions, and other genetic modifications. This makes NGS an ideal method for early-stage detection of sequence variants in drug development, where these variants might be present in minute quantities.
NGS methods do not detect sequence variants that arise from misincorporations, such as amino acid substitutions and amino acid modifications. It’s important to conduct sequence variant analysis at both the genomic and peptide level to understand the purity through both stages of recombinant expression.
Sequence Variant Analysis Service
Rapid Novor specializes in liquid chromatography-mass spectrometry (LC-MS) and offers a comprehensive suite of mass spectrometry-based characterization services for antibodies including:
- Sequence Variant Analysis: Detecting and characterizing even low-abundance sequence variants to ensure purity and consistency of biotherapeutics.
- Post-Translational Modification (PTM) Analysis: Identifying and quantifying modifications such as phosphorylation, isomerizations, and acetylation that can affect antibody function.
- Peptide Mapping: Providing detailed maps of protein sequences to confirm sequence identity, essential for quality assurance testing and regulatory compliance.
- Glycan Analysis: Characterizing glycan structures attached to antibodies, which are crucial for their biological activity and therapeutic efficacy.
- Intact Mass Analysis: Determining the molecular weight of intact antibodies to detect any mass shifts indicative of sequence variants or PTMs.
- Disulfide Bond Analysis: Identifying and confirming the disulfide bond linkages within antibodies to ensure proper folding and structural integrity.
Contact our scientists to learn more about antibody characterization and sequence variant analysis.
Talk to Our Scientists.
We Have Sequenced 10,000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and developed the first recombinant polyclonal antibody diagnostics.
Talk to Our Scientists.
We Have Sequenced 9000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables timely and reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and ran the first recombinant polyclonal antibody diagnostics

