 Written by María Gerpe, PhD
Written by María Gerpe, PhD
July 18, 2021
Introduction
Protein mass spectrometry refers to the use of mass spectrometry in the study and characterization of proteins, including their quantification, profiling, interaction mapping, and identification of their post-translational modifications (1,2). Protein mass spectrometry may also be referred to as mass spectrometry-based proteomics. Mass spectrometry-based proteomics consist of three approaches: top-down, middle-down, and bottom-up proteomics (2).
Top-down proteomics refers to mass spectrometry analysis of samples containing undigested or whole proteins (3,4). It’s typically executed with high resolution matrix-assisted laser desorption ionization (MALDI) mass spectrometers, but an orbitrap relying on electrospray ionization (ESI) may also be used (3,5,6). In contrast, bottom-up proteomics refers to the mass spectrometry-based study of protein fragments from protein digestions (7).
A sub-branch of bottom-up proteomics is shotgun proteomics, a widely used proteomics approach which attempts to identify proteins in complex mixtures through mass spectrometry and high performance liquid chromatography (7). Finally, middle-down proteomics started to gain traction circa 2012 as a way of compromising between top-down and bottom-up approaches. In the middle-down approach, larger peptide fragments are analyzed by the mass spectrometer (8).
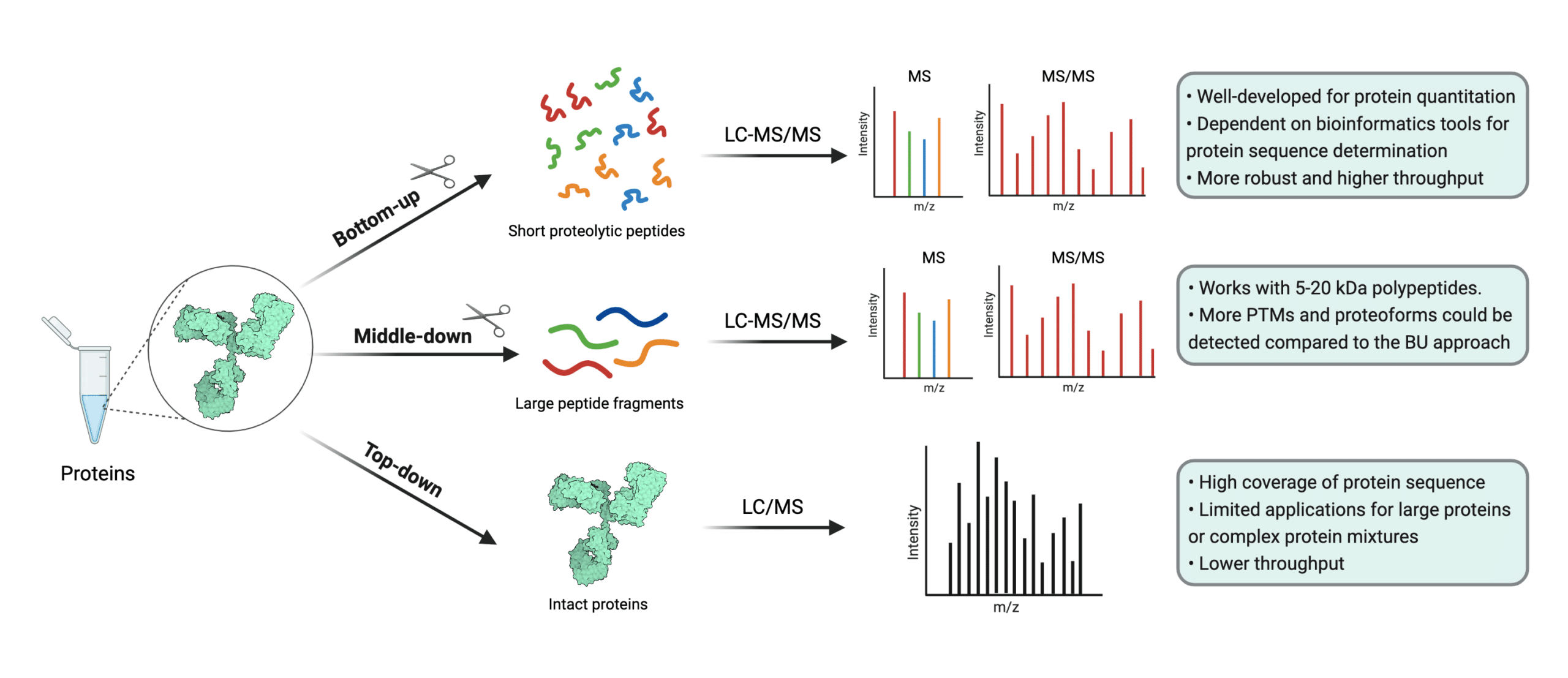
Figure 1. Illustration of the three approaches of mass spectrometry-based proteomics. Top-down methods often require MALDI instruments, but ESI instruments may be used.
Protein Mass Spectrometry at Work
A basic mass spectrometer contains an inlet that introduces digested or whole proteins to the mass spectrometer, where they are ionized at an ion source. In the case of MALDI spectrometers, a soft ionization technique is used in which a plate or matrix bearing the proteins is hit by a laser so they may move into the gas phase without fragmentation. Macromolecules may not withstand the heat and may not be amenable to MALDI-based protein mass spectrometry. In ESI-based protein mass spectrometers, the ion source is an electrospray (5,6).
Through the mass spectrometer, the flow of charged molecules is captured by ion detectors that provide feedback to an instrument control capable of adjusting the kinetic energy (MALDI instruments), and frequency and/or voltage (ESI instruments), and measuring the m/z ratio of the molecule.
ESI-based Protein Mass Spectrometry
In ESI-based protein mass spectrometry, the electrospray promotes ion flow toward mass analyzers. Mass analyzers may contain rods (quadrupole mass filter) or ring electrodes (ion trap) surrounding the flow of ions. The polarized rods or electrodes will attract peptide cations of a certain mass-to-charge (m/z) ratio to essentially ‘trap’ them; the ions that continually flow through will be detected by a sensor called the ion detector, where the ions will be measured based on their mass, charge and the time at which they have been recorded. An example of a mass analyzer is the quadrupole (5,6).
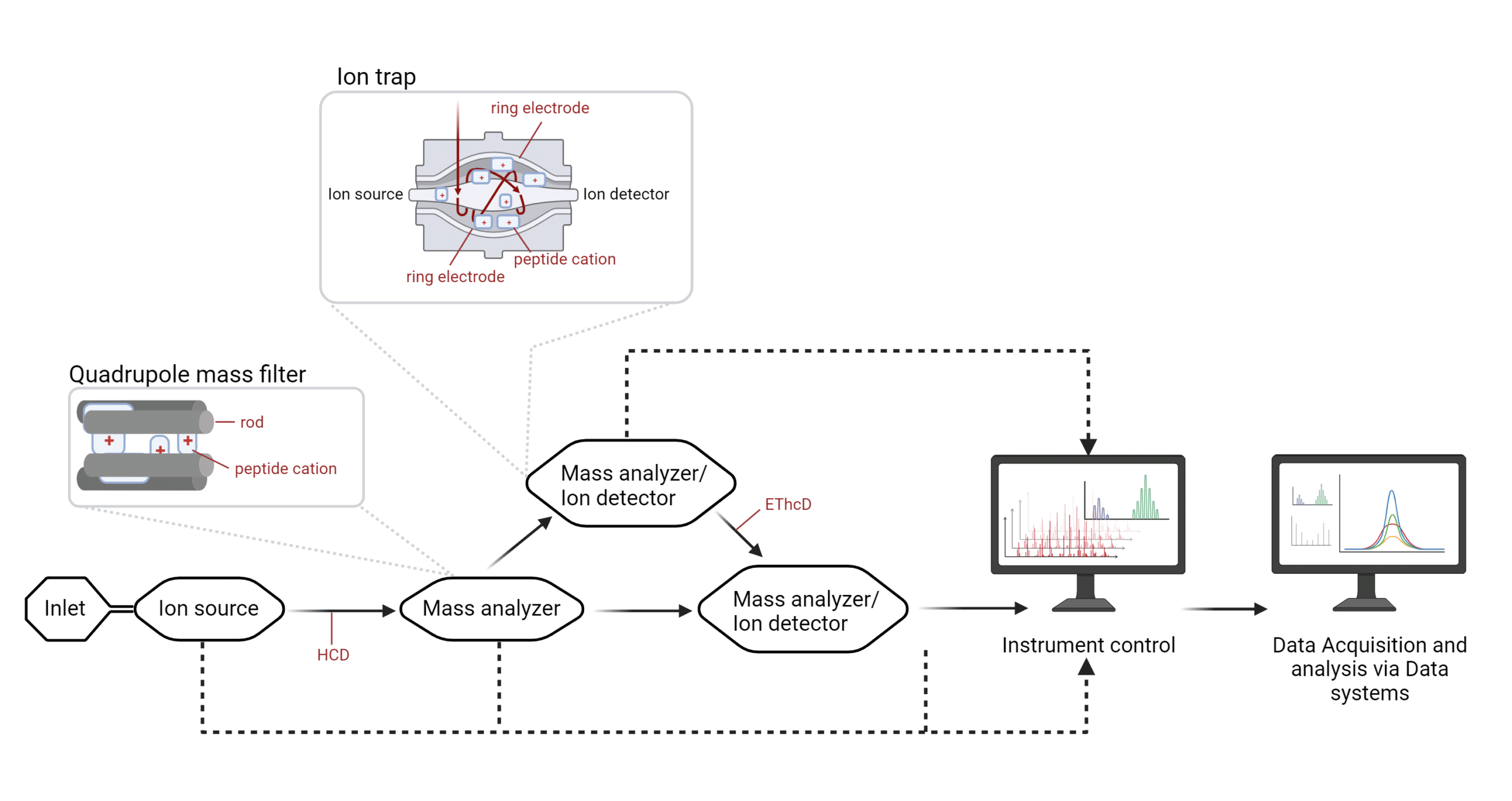
Figure 2. Infographic of an ESI-based mass spectrometer.
While ESI-based MS instruments initially contained one mass analyzer and one ion detector, nowadays, ion cells are typically flanked by the ion source and the ion sensor and sandwiched between electrodes. The latter can modulate the frequency and voltage to ‘select’ a window for the desired m/z ratio and then redirect the flow of ions for additional fragmentation. An example of an ion cell that can act as either, or both a mass analyzer and an ion detector is the ion trap. The ion source, and mass analyzers and ion detectors are all kept under vacuum.
As peptides travel between mass analyzers and ion detectors, collisions can fragment these peptides further via high-energy collision dissociation (HCD), or electron-transfer high-energy collision dissociation (EThcD). All of the internal systems feed into the instrument control. Essentially, at any of these points, data can be detected for the user may select a specific range of ions and record the spectra of these fragments for data analysis.
A mass spectrometer that comprises different sequential cells with mass analyzer and ion detector capabilities is referred to as a tandem mass spectrometer. Mass spectrometers used to only be able to house a mass analyzer and an ion detector. However, mass spectrometry technology has evolved to such a sophisticated level that current tandem mass spectrometers house many more components than original instruments. Whereas they used to be huge, with the advent of the benchtop mass spectrometer, these instruments can now conveniently fit in one corner of the laboratory (5).
Applications of Protein Mass Spectrometry
Because mass spectrometers are designed with a specific purpose in mind, the type of spectrometer is tied to specific applications. ESI-based mass spectrometers are especially suited to sequence analysis, whereas MALDI-based spectrometers are well suited for mass analysis. The following are common applications of protein mass spectrometry:
- Identification and verification of proteins (e.g., intact mass analysis, peptide mass fingerprinting or peptide mapping such as MATCHmAb™)
- Protein quantification (e.g., isotope labeling quantitative proteomics, non-labeled MALDI-based proteomics, and non-labeled, profiling ESI-based proteomics such as de novo antibody sequencing-based immune profiling)
- Protein structure determination (e.g., hydrogen deuterium exchange, or facilitated through de novo antibody protein sequencing)
- Determination of the protein sequence (e.g., proteogenomics, and protein sequencing by tandem mass spectrometry such as REmAb® and REpAb®)
Protein Mass Spectrometry at Rapid Novor
Rapid Novor’s offers antibody sequencing services and polyclonal sequencing services using LC-MS/MS. As of 2021, we are the leading protein sequencing company focusing on antibodies with over 3000 antibody and protein sequencing projects worldwide.
References
- B. Domon and R. Aebersold, “Mass Spectrometry and Protein Analysis,” Science, vol. 312, no. 5771, pp. 212-217, 2006.
- K. A. Resing and N. G. Ahn, “Proteomics strategies for protein identification,” FEBS Letters, vol. 579, no. 4, pp. 885-889, 2005.
- Catherman, AD, Skinner, OS, and Kelleher, NL. “Top Down Proteomics: Facts and Perspectives.” Biochem Biophys Res Commun. 445(4): 683–693. 2014
- D. P. Donnelly, C. . M. Rawlins, C. . J. DeHart, L. Fornelli, L. F. Schachner, Z. Lin, J. L. Lippens, K. C. Aluri, R. Sarin , B. Chen, C. Lantz , W. Jung , K. R. Johnson , A. Koller , J. J. Wolff , I. D. G. Campuzano, J. R. Auclair, A. R. Ivanov, J. P. Whitelegge, L. Paša-Tolić, J. Chamot-Rooke, P. O. Danis, L. M. Smith, Y. O. Tsybin, J. A. Loo, Y. Ge, N. L. Kelleher and J. N. Agar, “Best practices and benchmarks for intact protein analysis for top-down mass spectrometry,” Nature Methods, vol. 16, no. 7, pp. 587-594, 2019.
- M. Kinter and N. E. Sherman, “Ionization Methods,” in Protein Sequencing and Identification Using Tandem Mass Spectrometry, D. M. Desiderio and N. M. M. Nibbering, Eds., Toronto, Wiley-Interscience, 2000, p. 35.
- M. Kinter and N. E. Sherman, “Ionization Methods,” in Protein Sequencing and Identification Using Tandem Mass Spectrometry, D. M. Desiderio and N. M. M. Nibbering, Eds., Toronto, Wiley-Interscience, 2000, p. 35.
- Y. Zhang, B. R. Fonslow, B. Shan, M.-C. Baek and J. R. Yates, III, “Protein Analysis by Shotgun/Bottom-up Proteomics,” Chemical Reviews, vol. 113, no. 4, p. 2343–2394, 2014.
- C. Wu, J. C. Tran, L. Zamdborg, K. R. Durbin, M. Li, D. R. Ahlf, B. P. Early, P. M. Thomas, J. V. Sweedler and N. L. Kelleher, “A Protease for Middle Down Proteomics,” Nature Methods, vol. 9, no. 8, pp. 822-824, 2012.
Talk to Our Scientists.
We Have Sequenced 10,000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and developed the first recombinant polyclonal antibody diagnostics.
Talk to Our Scientists.
We Have Sequenced 9000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables timely and reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and ran the first recombinant polyclonal antibody diagnostics

